Regulatory divergence modifies limb length between mammals
- PMID: 18198333
- PMCID: PMC2192750
- DOI: 10.1101/gad.1620408
Regulatory divergence modifies limb length between mammals
Abstract
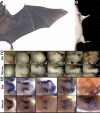
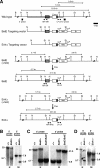
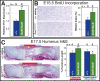
Natural selection acts on variation within populations, resulting in modified organ morphology, physiology, and ultimately the formation of new species. Although variation in orthologous proteins can contribute to these modifications, differences in DNA sequences regulating gene expression may be a primary source of variation. We replaced a limb-specific transcriptional enhancer of the mouse Prx1 locus with the orthologous sequence from a bat. Prx1 expression directed by the bat enhancer results in elevated transcript levels in developing forelimb bones and forelimbs that are significantly longer than controls because of endochondral bone formation alterations. Surprisingly, deletion of the mouse Prx1 limb enhancer results in normal forelimb length and Prx1 expression, revealing regulatory redundancy. These findings suggest that mutations accumulating in pre-existing noncoding regulatory sequences within a population are a source of variation for the evolution of morphological differences between species and that cis-regulatory redundancy may facilitate accumulation of such mutations.
Figures

Comment in
-
Understanding of bat wing evolution takes flight.Genes Dev. 2008 Jan 15;22(2):121-4. doi: 10.1101/gad.1639108. Genes Dev. 2008. PMID: 18198331 Free PMC article. No abstract available.
References
-
- Atchison M.L. Enhancers: Mechanisms of action and cell specificity. Annu. Rev. Cell Biol. 1988;4:127–153. - PubMed
-
- Ausubel F.M. Current protocols in molecular biology. John Wiley & Sons; New York: 1989.
-
- Behringer R.R., Finegold M.J., Cate R.L. Mullerian-inhibiting substance function during mammalian sexual development. Cell. 1994;79:415–425. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
