The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus
- PMID: 18198338
- PMCID: PMC2192755
- DOI: 10.1101/gad.1601808
The dynamic interplay between a cell fate determinant and a lysozyme homolog drives the asymmetric division cycle of Caulobacter crescentus
Abstract
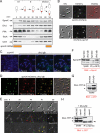
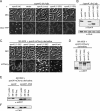
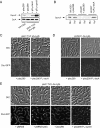
Caulobacter crescentus divides asymmetrically into a swarmer cell and a stalked cell, a process that is governed by the imbalance in phosphorylated levels of the DivK cell fate determinant in the two cellular compartments. The asymmetric polar localization of the DivJ kinase results in its specific inheritance in the stalked daughter cell where it phosphorylates DivK. The mechanism for the polar positioning of DivJ is poorly understood. SpmX, an uncharacterized lysozyme homolog, is shown here to control DivJ localization and activation. In the absence of SpmX, DivJ is delocalized and dysfunctional, resulting in developmental defects caused by an insufficiency in phospho-DivK. While SpmX stimulates DivK phosphorylation in the stalked cell, unphosphorylated DivK in the swarmer cell activates an intricate transcriptional cascade that leads to the production of the spmX message. This event primes the swarmer cell for the impending transition into a stalked cell, a transition that is sparked by the abrupt accumulation and localization of SpmX to the future stalked cell pole. Localized SpmX then recruits and stimulates DivJ, and the resulting phospho-DivK implements the stalked cell fate. The dynamic interplay between SpmX and DivK is at the heart of the molecular circuitry that sustains the Caulobacter developmental cycle.
Figures




References
-
- Alley M.R., Maddock J.R., Shapiro L. Polar localization of a bacterial chemoreceptor. Genes & Dev. 1992;6:825–836. - PubMed
-
- Biondi E.G., Reisinger S.J., Skerker J.M., Arif M., Perchuk B.S., Ryan K.R., Laub M.T. Regulation of the bacterial cell cycle by an integrated genetic circuit. Nature. 2006a;444:899–904. - PubMed
-
- Biondi E.G., Skerker J.M., Arif M., Prasol M.S., Perchuk B.S., Laub M.T. A phosphorelay system controls stalk biogenesis during cell cycle progression in Caulobacter crescentus. Mol. Microbiol. 2006b;59:386–401. - PubMed
-
- Chen J.C., Viollier P.H., Shapiro L. A membrane metalloprotease participates in the sequential degradation of a Caulobacter polarity determinant. Mol. Microbiol. 2005;55:1085–1103. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
