Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum
- PMID: 18199763
- PMCID: PMC6670353
- DOI: 10.1523/JNEUROSCI.2986-07.2008
Ctip2 controls the differentiation of medium spiny neurons and the establishment of the cellular architecture of the striatum
Abstract
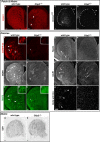
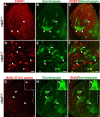
Striatal medium spiny neurons (MSN) are critically involved in motor control, and their degeneration is a principal component of Huntington's disease. We find that the transcription factor Ctip2 (also known as Bcl11b) is central to MSN differentiation and striatal development. Within the striatum, it is expressed by all MSN, although it is excluded from essentially all striatal interneurons. In the absence of Ctip2, MSN do not fully differentiate, as demonstrated by dramatically reduced expression of a large number of MSN markers, including DARPP-32, FOXP1, Chrm4, Reelin, MOR1 (mu-opioid receptor 1), glutamate receptor 1, and Plexin-D1. Furthermore, MSN fail to aggregate into patches, resulting in severely disrupted patch-matrix organization within the striatum. Finally, heterotopic cellular aggregates invade the Ctip2-/- striatum, suggesting a failure by MSN to repel these cells in the absence of Ctip2. This is associated with abnormal dopaminergic innervation of the mutant striatum and dramatic changes in gene expression, including dysregulation of molecules involved in cellular repulsion. Together, these data indicate that Ctip2 is a critical regulator of MSN differentiation, striatal patch development, and the establishment of the cellular architecture of the striatum.
Figures






References
-
- Albin RL, Young AB, Penney JB. The functional anatomy of basal ganglia disorders. Trends Neurosci. 1989;12:366–375. - PubMed
-
- Anderson KD, Reiner A. Immunohistochemical localization of DARPP-32 in striatal projection neurons and striatal interneurons: implications for the localization of D1-like dopamine receptors on different types of striatal neurons. Brain Res. 1991;568:235–243. - PubMed
-
- Anderson SA, Eisenstat DD, Shi L, Rubenstein JL. Interneuron migration from basal forebrain to neocortex: dependence on Dlx genes. Science. 1997a;278:474–476. - PubMed
-
- Anderson SA, Qiu M, Bulfone A, Eisenstat DD, Meneses J, Pedersen R, Rubenstein JL. Mutations of the homeobox genes Dlx-1 and Dlx-2 disrupt the striatal subventricular zone and differentiation of late born striatal neurons. Neuron. 1997b;19:27–37. - PubMed
-
- Arlotta P, Molyneaux BJ, Chen J, Inoue J, Kominami R, Macklis JD. Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron. 2005;45:207–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
