High-confidence prediction of global interactomes based on genome-wide coevolutionary networks
- PMID: 18199838
- PMCID: PMC2242690
- DOI: 10.1073/pnas.0709671105
High-confidence prediction of global interactomes based on genome-wide coevolutionary networks
Abstract
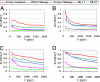
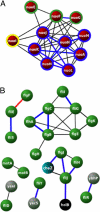
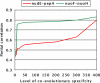
Interacting or functionally related protein families tend to have similar phylogenetic trees. Based on this observation, techniques have been developed to predict interaction partners. The observed degree of similarity between the phylogenetic trees of two proteins is the result of many different factors besides the actual interaction or functional relationship between them. Such factors influence the performance of interaction predictions. One aspect that can influence this similarity is related to the fact that a given protein interacts with many others, and hence it must adapt to all of them. Accordingly, the interaction or coadaptation signal within its tree is a composite of the influence of all of the interactors. Here, we introduce a new estimator of coevolution to overcome this and other problems. Instead of relying on the individual value of tree similarity between two proteins, we use the whole network of similarities between all of the pairs of proteins within a genome to reassess the similarity of that pair, thereby taking into account its coevolutionary context. We show that this approach offers a substantial improvement in interaction prediction performance, providing a degree of accuracy/coverage comparable with, or in some cases better than, that of experimental techniques. Moreover, important information on the structure, function, and evolution of macromolecular complexes can be inferred with this methodology.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- van Kesteren RE, Tensen CP, Smit AB, van Minnen J, Kolakowski LF, Meyerhof W, Richter D, van Heerikhuizen H, Vreugdenhil E, Geraerts WP. J Biol Chem. 1996;271:3619–3626. - PubMed
-
- Fryxell KJ. Trends Genet. 1996;12:364–369. - PubMed
-
- Goh C-S, Bogan AA, Joachimiak M, Walther D, Cohen FE. J Mol Biol. 2000;299:283–293. - PubMed
-
- Pazos F, Valencia A. Protein Eng. 2001;14:609–614. - PubMed
-
- Pazos F, Ranea JAG, Juan D, Sternberg MJE. J Mol Biol. 2005;352:1002–1015. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

