CpG methylation of an endogenous retroviral enhancer inhibits transcription factor binding and activity
- PMID: 1820217
- PMCID: PMC5952189
CpG methylation of an endogenous retroviral enhancer inhibits transcription factor binding and activity
Abstract
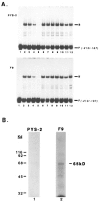
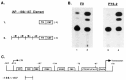
The endogenous retrovirus, intracisternal A-particle (IAP), is expressed at unique stages during murine embryogenesis and is also activated during the in vitro differentiation of F9 cells. We have examined the DNA elements and protein factors that control IAP expression during F9 differentiation. In the present study an IAP upstream enhancer (IUE) is identified by transient transfection assays and found to be active in both undifferentiated and differentiated cells. Further analyses reveal that a ubiquitous 65 kDa protein factor, the IUE binding protein (IUEB), binds with the IUE. Site-specific methylation within the IUEB binding site strongly inhibits both IUEB binding and IUE transcriptional activity, suggesting that methylation may regulate IUE function and IAP expression.
Figures




References
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., and Struhl K., eds. (1990), in Current Protocols in Molecular Biology, Green Publishing Associates and Wiley Interscience, John Wiley and Sons, New York.
-
- Becker P. B., Ruppert S., and Schutz G. (1987), Cell 51, 435–443. - PubMed
-
- Briggs M. R., Kadonaga J. T., Bell S. P., and Tjian R. (1986), Science 234, 47–52. - PubMed
-
- Burbelo P. D., Horikoshi S., and Yamada Y. (1990), J Biol Chem 265, 4839–4843. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
