Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae
- PMID: 18202364
- PMCID: PMC2206066
- DOI: 10.1534/genetics.107.081315
Identification of novel activation mechanisms for FLO11 regulation in Saccharomyces cerevisiae
Abstract
Adhesins play a central role in the cellular response of eukaryotic microorganisms to their host environment. In pathogens such as Candida spp. and other fungi, adhesins are responsible for adherence to mammalian tissues, and in Saccharomyces spp. yeasts also confer adherence to solid surfaces and to other yeast cells. The analysis of FLO11, the main adhesin identified in Saccharomyces cerevisiae, has revealed complex mechanisms, involving both genetic and epigenetic regulation, governing the expression of this critical gene. We designed a genomewide screen to identify new regulators of this pivotal adhesin in budding yeasts. We took advantage of a specific FLO11 allele that confers very high levels of FLO11 expression to wild "flor" strains of S. cerevisiae. We screened for mutants that abrogated the increased FLO11 expression of this allele using the loss of the characteristic fluffy-colony phenotype and a reporter plasmid containing GFP controlled by the same FLO11 promoter. Using this approach, we isolated several genes whose function was essential to maintain the expression of FLO11. In addition to previously characterized activators, we identified a number of novel FLO11 activators, which reveal the pH response pathway and chromatin-remodeling complexes as central elements involved in FLO11 activation.
Figures


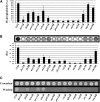
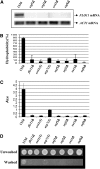


References
-
- Bayly, J. C., L. M. Douglas, I. S. Pretorius, F. F. Bauer and A. M. Dranginis, 2005. Characteristics of Flo11-dependent flocculation in Saccharomyces cerevisiae. FEMS Yeast Res. 5: 1151–1156. - PubMed
-
- Bjorklund, S., and C. M. Gustafsson, 2005. The yeast Mediator complex and its regulation. Trends Biochem. Sci. 30: 240–244. - PubMed
-
- Boukaba, A., E. I. Georgieva, F. A. Myers, A. W. Thorne, G. Lopez-Rodas et al., 2004. A short-range gradient of histone H3 acetylation and Tup1p redistribution at the promoter of the Saccharomyces cerevisiae SUC2 gene. J. Biol. Chem. 279: 7678–7684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

