Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation
- PMID: 18202370
- PMCID: PMC2206074
- DOI: 10.1534/genetics.107.083212
Inhibition of transcription by the Caenorhabditis elegans germline protein PIE-1: genetic evidence for distinct mechanisms targeting initiation and elongation
Abstract
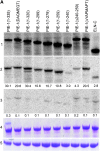
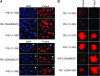
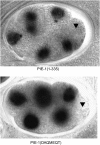
In Caenorhabditis elegans embryos, specification of the germ lineage depends on PIE-1, a maternal protein that blocks mRNA transcription in germline blastomeres. Studies in mammalian cell culture have suggested that PIE-1 inhibits P-TEFb, a kinase that phosphorylates serine 2 in the carboxyl-terminal domain (CTD) repeats of RNA polymerase II during transcriptional elongation. We have tested this hypothesis using an in vivo complementation assay for PIE-1 function. Our results support the view that PIE-1 inhibits P-TEFb using the CTD-like motif YAPMAPT. This activity is required to block serine 2 phosphorylation in germline blastomeres, but unexpectedly is not essential for transcriptional repression or specification of the germline. We find that sequences outside of the YAPMAPT are required to inhibit serine 5 phosphorylation, and that this second inhibitory mechanism is essential for transcriptional repression and specification of the germ lineage. Our results suggest that PIE-1 uses partially redundant mechanisms to block transcription by targeting both the initiation and elongation phases of the transcription cycle.
Figures

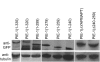

References
-
- D'Agostino, I., C. Merritt, P. L. Chen, G. Seydoux and K. Subramaniam, 2006. Translational repression restricts expression of the C. elegans Nanos homolog NOS-2 to the embryonic germline. Dev. Biol. 292: 244–252. - PubMed
-
- Deshpande, G., G. Calhoun and P. Schedl, 2004. Overlapping mechanisms function to establish transcriptional quiescence in the embryonic Drosophila germline. Development 131: 1247–1257. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

