An unstable targeted allele of the mouse Mitf gene with a high somatic and germline reversion rate
- PMID: 18202372
- PMCID: PMC2206076
- DOI: 10.1534/genetics.107.081893
An unstable targeted allele of the mouse Mitf gene with a high somatic and germline reversion rate
Abstract
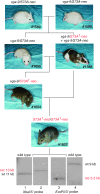
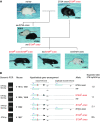
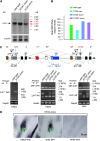
The mouse Mitf gene encodes a transcription factor that is regulated by serine phosphorylation and is critical for the development of melanin-containing pigment cells. To test the role of phosphorylation at a particular serine, S73 in exon 2 of Mitf, we used a standard targeting strategy in mouse embryonic stem cells to change the corresponding codon into one encoding an alanine. By chance, we generated an allele in which 85,222 bp of wild-type Mitf sequence are duplicated and inserted into an otherwise correctly targeted Mitf gene. Depending on the presence or absence of a neomycin resistance cassette, this genomic rearrangement leads to animals with a white coat with or without pigmented spots or a gray coat with obligatory white and black spots. Several independent, genetically stable germline revertants that lacked the duplicated wild-type sequence but retained the targeted codon were then derived. These animals were normally pigmented, indicating that the serine-to-alanine mutation is not deleterious to melanocyte development. The fact that mosaic coat reversions occur in all mice lacking the neo-cassette and that approximately 1% of these transmit a reverted allele to their offspring places this mutation among those with the highest spontaneous reversion rates in mammals.
Figures




References
-
- Arnheiter, H., L. Hou, M.-T. T. Nguyen, K. Bismuth, T. Csermely et al., 2006. Mitf—A Matter of Life and Death for the Developing Melanocyte. Humana Press, Totowa, NJ.
-
- Bennett, D. C., and M. L. Lamoreux, 2003. The color loci of mice: a genetic century. Pigment Cell Res. 16: 333–344. - PubMed
-
- Brilliant, M. H., Y. Gondo and E. M. Eicher, 1991. Direct molecular identification of the mouse pink-eyed unstable mutation by genome scanning. Science 252: 566–569. - PubMed
-
- Conrad, B., and S. E. Antonarakis, 2007. Gene duplication: a drive for phenotypic diversity and cause of human disease. Annu. Rev. Genomics Hum. Genet. 8: 17–35. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

