Testing for archaic hominin admixture on the X chromosome: model likelihoods for the modern human RRM2P4 region from summaries of genealogical topology under the structured coalescent
- PMID: 18202385
- PMCID: PMC2206091
- DOI: 10.1534/genetics.107.080432
Testing for archaic hominin admixture on the X chromosome: model likelihoods for the modern human RRM2P4 region from summaries of genealogical topology under the structured coalescent
Abstract
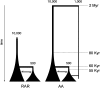
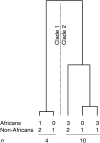
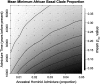
A 2.4-kb stretch within the RRM2P4 region of the X chromosome, previously sequenced in a sample of 41 globally distributed humans, displayed both an ancient time to the most recent common ancestor (e.g., a TMRCA of approximately 2 million years) and a basal clade composed entirely of Asian sequences. This pattern was interpreted to reflect a history of introgressive hybridization from archaic hominins (most likely Asian Homo erectus) into the anatomically modern human genome. Here, we address this hypothesis by resequencing the 2.4-kb RRM2P4 region in 131 African and 122 non-African individuals and by extending the length of sequence in a window of 16.5 kb encompassing the RRM2P4 pseudogene in a subset of 90 individuals. We find that both the ancient TMRCA and the skew in non-African representation in one of the basal clades are essentially limited to the central 2.4-kb region. We define a new summary statistic called the minimum clade proportion (pmc), which quantifies the proportion of individuals from a specified geographic region in each of the two basal clades of a binary gene tree, and then employ coalescent simulations to assess the likelihood of the observed central RRM2P4 genealogy under two alternative views of human evolutionary history: recent African replacement (RAR) and archaic admixture (AA). A molecular-clock-based TMRCA estimate of 2.33 million years is a statistical outlier under the RAR model; however, the large variance associated with this estimate makes it difficult to distinguish the predictions of the human origins models tested here. The pmc summary statistic, which has improved power with larger samples of chromosomes, yields values that are significantly unlikely under the RAR model and fit expectations better under a range of archaic admixture scenarios.
Figures



References
-
- Anton, S. C., and C. C. Swisher, 2004. Early dispersal of Homo from Africa. Annu. Rev. Anthropol. 33: 271–296.
-
- Basu, A., and P. Majumdar, 2003. A comparison of two popular statistical methods for estimating the time to most recent common ancestor (TMRCA) from a sample of DNA sequences. J. Genet. 82: 7–12. - PubMed
-
- Cann, H. M., C. de Toma, L. Cazes, M. F. Legrand, V. Morel et al., 2002. A human genome diversity cell line panel. Science 296: 261–262. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

