Accuracy of genomic selection using different methods to define haplotypes
- PMID: 18202394
- PMCID: PMC2206101
- DOI: 10.1534/genetics.107.080838
Accuracy of genomic selection using different methods to define haplotypes
Abstract
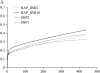
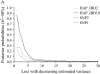
Genomic selection uses total breeding values for juvenile animals, predicted from a large number of estimated marker haplotype effects across the whole genome. In this study the accuracy of predicting breeding values is compared for four different models including a large number of markers, at different marker densities for traits with heritabilities of 50 and 10%. The models estimated the effect of (1) each single-marker allele [single-nucleotide polymorphism (SNP)1], (2) haplotypes constructed from two adjacent marker alleles (SNP2), and (3) haplotypes constructed from 2 or 10 markers, including the covariance between haplotypes by combining linkage disequilibrium and linkage analysis (HAP_IBD2 and HAP_IBD10). Between 119 and 2343 polymorphic SNPs were simulated on a 3-M genome. For the trait with a heritability of 10%, the differences between models were small and none of them yielded the highest accuracies across all marker densities. For the trait with a heritability of 50%, the HAP_IBD10 model yielded the highest accuracies of estimated total breeding values for juvenile and phenotyped animals at all marker densities. It was concluded that genomic selection is considerably more accurate than traditional selection, especially for a low-heritability trait.
Figures







References
-
- Falconer, D. S., and T. F. C. Mackay, 1996. Introduction to Quantitative Genetics. Longman Group, Essex, UK.
-
- Fernando, R. L., and M. Grossman, 1989. Marker assisted selection using best linear unbiased prediction. Genet. Sel. Evol. 21: 467–477.
-
- Hayes, B. J., A. J. Chamberlain and M. E. Goddard, 2006. Use of markers in linkage disequilibrium with QTL in breeding programs. Proceedings of the 8th World Congress on Genetics Applied to Livestock Production, Belo Horizonte, MG, Brazil, Communication 30–06.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

