Transcription regulation of the type II restriction-modification system AhdI
- PMID: 18203750
- PMCID: PMC2275141
- DOI: 10.1093/nar/gkm1116
Transcription regulation of the type II restriction-modification system AhdI
Abstract
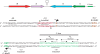
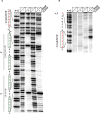
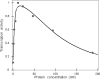
The Restriction-modification system AhdI contains two convergent transcription units, one with genes encoding methyltransferase subunits M and S and another with genes encoding the controller (C) protein and the restriction endonuclease (R). We show that AhdI transcription is controlled by two independent regulatory loops that are well-optimized to ensure successful establishment in a naïve bacterial host. Transcription from the strong MS promoter is attenuated by methylation of an AhdI site overlapping the -10 element of the promoter. Transcription from the weak CR promoter is regulated by the C protein interaction with two DNA-binding sites. The interaction with the promoter-distal high-affinity site activates transcription, while interaction with the weaker promoter-proximal site represses it. Because of high levels of cooperativity, both C protein-binding sites are always occupied in the absence of RNA polymerase, raising a question how activated transcription is achieved. We develop a mathematical model that is in quantitative agreement with the experiment and indicates that RNA polymerase outcompetes C protein from the promoter-proximal-binding site. Such an unusual mechanism leads to a very inefficient activation of the R gene transcription, which presumably helps control the level of the endonuclease in the cell.
Figures





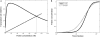
References
-
- Jeltsch A, Pingoud A. Horizontal gene transfer contributes to the wide distribution and evolution of type II restriction-modification systems. J. Mol. Evol. 1996;42:91–96. - PubMed
-
- Anton BP, Heiter DF, Benner JS, Hess EJ, Greenough L, Moran LS, Slatko BE, Brooks JE. Cloning and characterization of the BglII restriction-modification system reveals a possible evolutionary footprint. Gene. 1997;187:19–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

