BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP
- PMID: 18203820
- PMCID: PMC2234109
- DOI: 10.1073/pnas.0702132105
BiP mutants that are unable to interact with endoplasmic reticulum DnaJ proteins provide insights into interdomain interactions in BiP
Abstract
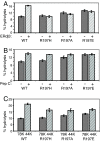
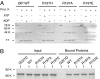
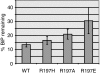
The heat shock protein (Hsp)70 family of molecular chaperones interacts with unfolded proteins through a C-terminal substrate-binding domain (SBD) that is controlled by nucleotide binding to the N-terminal domain. The ATPase cycle is regulated by cochaperones, including DnaJ proteins that accelerate ATP hydrolysis to stabilize the Hsp70-substrate complex. We found that R197 in hamster BiP, which resides at the surface of the nucleotide-binding domain, is critical for both association with endoplasmic reticulum DnaJ proteins and interaction with the SBD. Decreasing the positive charge at this residue enhanced basal ATPase activity, destabilized interaction with the SBD, and reduced substrate release both in vitro and in vivo. Mutation of three glutamic acids in the SBD mimicked many of these effects. Our data provide insights into communications between the two domains and suggest a mechanism by which DnaJ proteins increase ATP hydrolysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

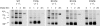

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

