Bacterial communities of Bartonella-positive fleas: diversity and community assembly patterns
- PMID: 18203862
- PMCID: PMC2258626
- DOI: 10.1128/AEM.02090-07
Bacterial communities of Bartonella-positive fleas: diversity and community assembly patterns
Abstract
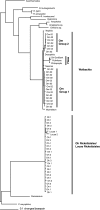
We investigated the bacterial communities of nine Bartonella-positive fleas (n = 6 Oropsylla hirsuta fleas and n = 3 Oropsylla montana fleas), using universal primers, clone libraries, and DNA sequencing. DNA sequences were used to classify bacteria detected in a phylogenetic context, to explore community assembly patterns within individual fleas, and to survey diversity patterns in dominant lineages.
Figures



References
-
- Furman, D. P. a. E. P. C. 1982. Order Siphonoptera, p. 207. In Manual of medical entomology. Cambridge University Press, Cambridge, United Kingdom.
-
- Gorham, C. H., Q. Q. Fang, and L. A. Durden. 2003. Wolbachia endosymbionts in fleas (Siphonaptera). J. Parasitol. 89:283-289. - PubMed
-
- Hubbard, C. A. 1947. Fleas of western North America. Hafner Publishing Company, New York, NY.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

