The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression
- PMID: 18206973
- PMCID: PMC2254522
- DOI: 10.1016/j.molcel.2007.12.015
The putative cancer stem cell marker USP22 is a subunit of the human SAGA complex required for activated transcription and cell-cycle progression
Abstract
Polycomb genes encode critical regulators of both normal stem cells and cancer stem cells. A gene signature that includes Polycomb genes and additional genes coregulated with Polycomb genes was recently identified. The expression of this signature has been reported to identify tumors with the cancer stem cell phenotypes of aggressive growth, metastasis, and therapy resistance. Most members of this 11 gene signature encode proteins with well-defined roles in human cancer. However, the function of the signature member USP22 remains unknown. We report that USP22 is a previously uncharacterized subunit of the human SAGA transcriptional cofactor complex. Within SAGA, USP22 deubiquitylates histone H2B. Furthermore, USP22 is recruited to specific genes by activators such as the Myc oncoprotein, where it is required for transcription. In support of a functional role within the Polycomb/cancer stem cell signature, USP22 is required for appropriate progression through the cell cycle.
Figures
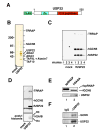
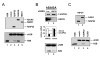
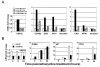
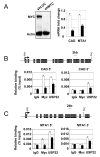

Comment in
-
Dubbing SAGA unveils new epigenetic crosstalk.Mol Cell. 2008 Feb 1;29(2):152-4. doi: 10.1016/j.molcel.2008.01.007. Mol Cell. 2008. PMID: 18243109
References
-
- Barlev NA, Liu L, Chehab NH, Mansfield K, Harris KG, Halazonetis TD, Berger SL. Acetylation of p53 activates transcription through recruitment of coactivators/histone acetyltransferases. Mol Cell. 2001;8:1243–1254. - PubMed
-
- Ben-Saadon R, Zaaroor D, Ziv T, Ciechanover A. The polycomb protein Ring1B generates self atypical mixed ubiquitin chains required for its in vitro histone H2A ligase activity. Mol Cell. 2006;24:701–711. - PubMed
-
- Blagosklonny MV. Target for cancer therapy: proliferating cells or stem cells. Leukemia. 2006;20:385–391. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

