Mechanisms of base selection by the Escherichia coli mispaired uracil glycosylase
- PMID: 18208817
- PMCID: PMC2276368
- DOI: 10.1074/jbc.M707174200
Mechanisms of base selection by the Escherichia coli mispaired uracil glycosylase
Abstract
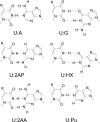
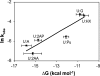
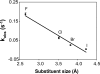
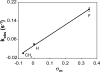
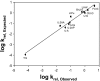
The repair of the multitude of single-base lesions formed daily in cells of all living organisms is accomplished primarily by the base excision repair pathway that initiates repair through a series of lesion-selective glycosylases. In this article, single-turnover kinetics have been measured on a series of oligonucleotide substrates containing both uracil and purine analogs for the Escherichia coli mispaired uracil glycosylase (MUG). The relative rates of glycosylase cleavage have been correlated with the free energy of helix formation and with the size and electronic inductive properties of a series of uracil 5-substituents. Data are presented that MUG can exploit the reduced thermodynamic stability of mispairs to distinguish U:A from U:G pairs. Discrimination against the removal of thymine results primarily from the electron-donating property of the thymine 5-methyl substituent, whereas the size of the methyl group relative to a hydrogen atom is a secondary factor. A series of parameters have been obtained that allow prediction of relative MUG cleavage rates that correlate well with observed relative rates that vary over 5 orders of magnitude for the series of base analogs examined. We propose that these parameters may be common among DNA glycosylases; however, specific glycosylases may focus more or less on each of the parameters identified. The presence of a series of glycosylases that focus on different lesion properties, all coexisting within the same cell, would provide a robust and partially redundant repair system necessary for the maintenance of the genome.
Figures
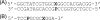

References
-
- Lindahl, T. (2000) Mutat. Res. 462 129–135 - PubMed
-
- Krokan, H. E., Nilsen, H., Skorpen, F., Otterlei, M., and Slupphaug, G. (2000) FEBS Lett. 476 73–77 - PubMed
-
- Sung, J. S., and Demple, B. (2006) FEBS J. 273 1620–1629 - PubMed
-
- Wilson, D. M., and Bohr, V. A. (2007) DNA Repair 6 544–559 - PubMed
-
- Saul, R. L., and Ames, B. N. (1986) Basic Life Sci. 38 529–535 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

