Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus
- PMID: 18209073
- PMCID: PMC3373268
- DOI: 10.4049/jimmunol.180.3.1758
Healthy human subjects have CD4+ T cells directed against H5N1 influenza virus
Abstract
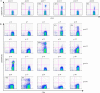
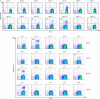
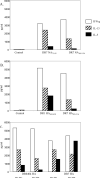
It is commonly perceived that the human immune system is naive to the newly emerged H5N1 virus. In contrast, most adults have been exposed to influenza A H1N1 and H3N2 viruses through vaccination or infection. Adults born before 1968 have likely been exposed to H2N2 viruses. We hypothesized that CD4(+) T cells generated in response to H1N1, H3N2, and H2N2 influenza A viruses also recognize H5N1 epitopes. Tetramer-guided epitope mapping and Ag-specific class II tetramers were used to identify H5N1-specific T cell epitopes and detect H5N1-specific T cell responses. Fifteen of 15 healthy subjects tested had robust CD4(+) T cell responses against matrix protein, nucleoprotein, and neuraminidase of the influenza A/Viet Nam/1203/2004 (H5N1) virus. These results are not surprising, because the matrix protein and nucleoprotein of influenza A viruses are conserved while the neuraminidase of the H5N1 virus is of the same subtype as that of the circulating H1N1 influenza strain. However, H5N1 hemagglutinin-reactive CD4(+) T cells were also detected in 14 of 14 subjects examined despite the fact that hemagglutinin is less conserved. Most were cross-reactive to H1, H2, or H3 hemagglutinin epitopes. H5N1-reactive T cells were also detected ex vivo, exhibited a memory phenotype, and were capable of secreting IFN-gamma, TNF-alpha, IL-5, and IL-13. These data demonstrate the presence of H5N1 cross-reactive T cells in healthy Caucasian subjects, implying that exposure to influenza A H1N1, H3N2, or H2N2 viruses through either vaccination or infection may provide partial immunity to the H5N1 virus.
Figures






References
-
- Olsen B, Munster VJ, Wallensten A, Waldenstrom J, Osterhaus AD, Fouchier RA. Global patterns of influenza a virus in wild birds. Science. 2006;312:384–388. - PubMed
-
- Beigel JH, Farrar J, Han AM, Hayden FG, Hyer R, de Jong MD, Lochindarat S, Nguyen TK, Nguyen TH, Tran TH, Nicoll A, Touch S, Yuen KY. Avian influenza A (H5N1) infection in humans. N. Engl. J. Med. 2005;353:1374–1385. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

