The comparative analysis of statistics, based on the likelihood ratio criterion, in the automated annotation problem
- PMID: 18211675
- PMCID: PMC2267706
- DOI: 10.1186/1471-2105-9-31
The comparative analysis of statistics, based on the likelihood ratio criterion, in the automated annotation problem
Abstract
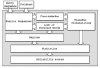
Background: This paper discusses the problem of automated annotation. It is a continuation of the previous work on the A4-algorithm (Adaptive algorithm of automated annotation) developed by Leontovich and others.
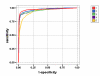
Results: A number of new statistics for the automated annotation of biological sequences is introduced. All these statistics are based on the likelihood ratio criterion.
Conclusion: Some of the statistics yield a prediction quality that is significantly higher (up to 1.5 times higher) in comparison with the results obtained with the A4-procedure.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

