Quantitative 1H NMR metabolomics reveals extensive metabolic reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures
- PMID: 18211706
- PMCID: PMC2257952
- DOI: 10.1186/1471-2229-8-5
Quantitative 1H NMR metabolomics reveals extensive metabolic reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures
Abstract
Background: Opium poppy (Papaver somniferum) produces a diverse array of bioactive benzylisoquinoline alkaloids and has emerged as a model system to study plant alkaloid metabolism. The plant is cultivated as the only commercial source of the narcotic analgesics morphine and codeine, but also produces many other alkaloids including the antimicrobial agent sanguinarine. Modulations in plant secondary metabolism as a result of environmental perturbations are often associated with the altered regulation of other metabolic pathways. As a key component of our functional genomics platform for opium poppy we have used proton nuclear magnetic resonance (1H NMR) metabolomics to investigate the interplay between primary and secondary metabolism in cultured opium poppy cells treated with a fungal elicitor.
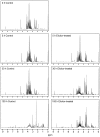
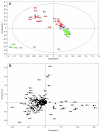
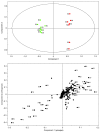
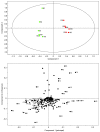
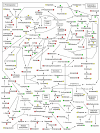
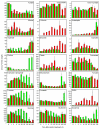
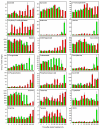
Results: Metabolite fingerprinting and compound-specific profiling showed the extensive reprogramming of primary metabolic pathways in association with the induction of alkaloid biosynthesis in response to elicitor treatment. Using Chenomx NMR Suite v. 4.6, a software package capable of identifying and quantifying individual compounds based on their respective signature spectra, the levels of 42 diverse metabolites were monitored over a 100-hour time course in control and elicitor-treated opium poppy cell cultures. Overall, detectable and dynamic changes in the metabolome of elicitor-treated cells, especially in cellular pools of carbohydrates, organic acids and non-protein amino acids were detected within 5 hours after elicitor treatment. The metabolome of control cultures also showed substantial modulations 80 hours after the start of the time course, particularly in the levels of amino acids and phospholipid pathway intermediates. Specific flux modulations were detected throughout primary metabolism, including glycolysis, the tricarboxylic acid cycle, nitrogen assimilation, phospholipid/fatty acid synthesis and the shikimate pathway, all of which generate secondary metabolic precursors.
Conclusion: The response of cell cultures to elicitor treatment involves the extensive reprogramming of primary and secondary metabolism, and associated cofactor biosynthetic pathways. A high-resolution map of the extensive reprogramming of primary and secondary metabolism in elicitor-treated opium poppy cell cultures is provided.
Figures

References
-
- Samanani N, Liscombe DK, Facchini PJ. Molecular cloning and characterization of norcoclaurine synthase, an enzyme catalyzing the first committed step in benzylisoquinoline alkaloid biosynthesis. Plant J. 2004;40:302–313. - PubMed
-
- Liscombe DK, MacLeod BP, Loukanina N, Nandi O, Facchini PJ. Evidence for the monophyletic evolution of benzylisoquinoline alkaloid biosynthesis in angiosperms. Phytochemistry. 2005;66:1374–1393. - PubMed
-
- Facchini PJ. Regulation of alkaloid biosynthesis in plants. Alkaloids Chem Biol. 2006;63:1–44. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

