Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells
- PMID: 18211900
- PMCID: PMC2293288
- DOI: 10.1074/jbc.M709446200
Alterations of BRMS1-ARID4A interaction modify gene expression but still suppress metastasis in human breast cancer cells
Abstract
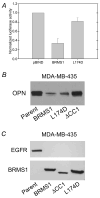
The BRMS1 metastasis suppressor interacts with the protein AT-rich interactive domain 4A (ARID4A, RBBP1) as part of SIN3.histone deacetylase chromatin remodeling complexes. These transcriptional co-repressors regulate diverse cell phenotypes depending upon complex composition. To define BRMS1 complexes and their roles in metastasis suppression, we generated BRMS1 mutants (BRMS1(mut)) and mapped ARID4A interactions. BRMS1(L174D) disrupted direct interaction with ARID4A in yeast two-hybrid genetic screens but retained an indirect association with ARID4A in MDA-MB-231 and -435 human breast cancer cell lines by co-immunoprecipitation. Deletion of the first coiled-coil domain (BRMS1(DeltaCC1)) did not disrupt direct interaction in yeast two-hybrid screens but did prevent association by co-immunoprecipitation. These results suggest altered complex composition with BRMS1(mut). Although basal transcription repression was impaired and the pro-metastatic protein osteopontin was differentially down-regulated by BRMS1(L174D) and BRMS1(DeltaCC1), both down-regulated the epidermal growth factor receptor and suppressed metastasis in MDA-MB-231 and -435 breast cancer xenograft models. We conclude that BRMS1(mut), which modifies the composition of a SIN3.histone deacetylase chromatin remodeling complex, leads to altered gene expression profiles. Because metastasis requires the coordinate expression of multiple genes, down-regulation of at least one important gene, such as the epidermal growth factor receptor, had the ability to suppress metastasis. Understanding which interactions are necessary for particular biochemical/cellular functions may prove important for future strategies targeting metastasis.
Figures



Similar articles
-
Over-expression of the BRMS1 family member SUDS3 does not suppress metastasis of human cancer cells.Cancer Lett. 2009 Apr 8;276(1):32-7. doi: 10.1016/j.canlet.2008.10.024. Epub 2008 Dec 13. Cancer Lett. 2009. PMID: 19070953 Free PMC article.
-
Breast cancer metastasis suppressor 1 (BRMS1) forms complexes with retinoblastoma-binding protein 1 (RBP1) and the mSin3 histone deacetylase complex and represses transcription.J Biol Chem. 2004 Jan 9;279(2):1562-9. doi: 10.1074/jbc.M307969200. Epub 2003 Oct 26. J Biol Chem. 2004. PMID: 14581478
-
The C-terminal putative nuclear localization sequence of breast cancer metastasis suppressor 1, BRMS1, is necessary for metastasis suppression.PLoS One. 2013;8(2):e55966. doi: 10.1371/journal.pone.0055966. Epub 2013 Feb 4. PLoS One. 2013. PMID: 23390556 Free PMC article.
-
Metastasis suppression by BRMS1 associated with SIN3 chromatin remodeling complexes.Cancer Metastasis Rev. 2012 Dec;31(3-4):641-51. doi: 10.1007/s10555-012-9363-y. Cancer Metastasis Rev. 2012. PMID: 22678236 Review.
-
Breast cancer metastasis suppressor 1: update.Clin Exp Metastasis. 2003;20(1):45-50. doi: 10.1023/a:1022542519586. Clin Exp Metastasis. 2003. PMID: 12650606 Review.
Cited by
-
Globular adiponectin enhances invasion in human breast cancer cells.Oncol Lett. 2016 Jan;11(1):633-641. doi: 10.3892/ol.2015.3965. Epub 2015 Nov 24. Oncol Lett. 2016. PMID: 26870258 Free PMC article.
-
Multiple forms of BRMS1 are differentially expressed in the MCF10 isogenic breast cancer progression model.Clin Exp Metastasis. 2009;26(2):89-96. doi: 10.1007/s10585-008-9216-9. Epub 2008 Oct 8. Clin Exp Metastasis. 2009. PMID: 18841483 Free PMC article.
-
BRMS1 coordinates with LSD1 and suppresses breast cancer cell metastasis.Am J Cancer Res. 2018 Oct 1;8(10):2030-2045. eCollection 2018. Am J Cancer Res. 2018. PMID: 30416854 Free PMC article.
-
Location, location, location: the BRMS1 protein and melanoma progression.BMC Med. 2012 Feb 22;10:19. doi: 10.1186/1741-7015-10-19. BMC Med. 2012. PMID: 22356729 Free PMC article. Review.
-
Whole-Exome Sequencing Reveals New Potential Mutations Genes for Primary Mucosa-Associated Lymphoid Tissue Lymphoma Arising From the Kidney.Front Oncol. 2021 Jan 8;10:609839. doi: 10.3389/fonc.2020.609839. eCollection 2020. Front Oncol. 2021. PMID: 33585230 Free PMC article.
References
-
- Langley RR, Fidler IJ. Endocrine Rev. 2007;28:297–321. - PubMed
-
- Nguyen DX, Massagué J. Nature Rev Genet. 2007;8:341–352. - PubMed
-
- Seraj MJ, Samant RS, Verderame MF, Welch DR. Cancer Res. 2000;60:2764–2769. - PubMed
-
- Samant RS, Seraj MJ, Saunders MM, Sakamaki T, Shevde LA, Harms JF, Leonard TO, Goldberg SF, Budgeon LR, Meehan WJ, Winter CR, Christensen ND, Verderame MF, Donahue HJ, Welch DR. Clin Exptl Metastasis. 2001;18:683–693. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

