Single particle image reconstruction of the human recombinant Kv2.1 channel
- PMID: 18212012
- PMCID: PMC2257891
- DOI: 10.1529/biophysj.107.118562
Single particle image reconstruction of the human recombinant Kv2.1 channel
Abstract
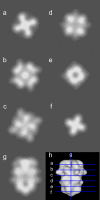
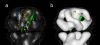
Kv2.1 channels are widely expressed in neuronal and endocrine cells and generate slowly activating K+ currents, which contribute to repolarization in these cells. Kv2.1 is expressed at high levels in the mammalian brain and is a major component of the delayed rectifier current in the hippocampus. In addition, Kv2.1 channels have been implicated in the regulation of membrane repolarization, cytoplasmic calcium levels, and insulin secretion in pancreatic beta-cells. They are therefore an important drug target for the treatment of Type II diabetes mellitus. We used electron microscopy and single particle image analysis to derive a three-dimensional density map of recombinant human Kv2.1. The tetrameric channel is egg-shaped with a diameter of approximately 80 A and a long axis of approximately 120 A. Comparison to known crystal structures of homologous domains allowed us to infer the location of the cytoplasmic and transmembrane assemblies. There is a very good fit of the Kv1.2 crystal structure to the assigned transmembrane assembly of Kv2.1. In other low-resolution maps of K+ channels, the cytoplasmic N-terminal and transmembrane domains form separate rings of density. In contrast, Kv2.1 displays contiguous density that connects the rings, such that there are no large windows between the channel interior and the cytoplasmic space. The crystal structure of KcsA is thought to be in a closed conformation, and the good fit of the KcsA crystal structure to the Kv2.1 map suggests that our preparations of Kv2.1 may also represent a closed conformation. Substantial cytoplasmic density is closely associated with the T1 tetramerization domain and is ascribed to the approximately 184 kDa C-terminal regulatory domains within each tetramer.
Figures






References
-
- Minor, D. L., Jr. 2001. Potassium channels: Life in the post-structural world. Curr. Opin. Struct. Biol. 11:408–414. - PubMed
-
- Yellen, G. 2002. The voltage-gated potassium channels and their relatives. Nature. 419:35–42. - PubMed
-
- Doyle, D. A., J. Morais Cabral, R. A. Pfuetzner, A. Kuo, J. M. Gulbis, S. L. Cohen, B. T. Chait, and R. MacKinnon. 1998. The structure of the potassium channel: Molecular basis of K+ conduction and selectivity. Science. 280:69–77. - PubMed
-
- Zhou, Y., J. H. Morais-Cabral, A. Kaufman, and R. MacKinnon. 2001. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature. 414:43–48. - PubMed
-
- Jiang, Y., A. Lee, J. Chen, V. Ruta, M. Cadene, B. T. Chait, and R. MacKinnon. 2003. X-ray structure of a voltage-dependent K+ channel. Nature. 423:33–41. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

