Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants
- PMID: 18212047
- PMCID: PMC2268442
- DOI: 10.1128/MCB.01755-07
Eaf1 is the platform for NuA4 molecular assembly that evolutionarily links chromatin acetylation to ATP-dependent exchange of histone H2A variants
Abstract
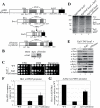
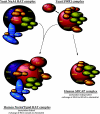
Eaf1 (for Esa1-associated factor 1) and Eaf2 have been identified as stable subunits of NuA4, a yeast histone H4/H2A acetyltransferase complex implicated in gene regulation and DNA repair. While both SWI3-ADA2-N-CoR-TF IIIB domain-containing proteins are required for normal cell cycle progression, their depletion does not affect the global Esa1-dependent acetylation of histones. In contrast to all other subunits, Eaf1 is found exclusively associated with the NuA4 complex in vivo. It serves as a platform that coordinates the assembly of functional groups of subunits into the native NuA4 complex. Eaf1 shows structural similarities with human p400/Domino, a subunit of the NuA4-related TIP60 complex. On the other hand, p400 also possesses an SWI2/SNF2 family ATPase domain that is absent from the yeast NuA4 complex. This domain is highly related to the yeast Swr1 protein, which is responsible for the incorporation of histone variant H2AZ in chromatin. Since all of the components of the TIP60 complex are homologous to SWR1 or NuA4 subunits, we proposed that the human complex corresponds to a physical merge of two yeast complexes. p400 function in TIP60 then would be accomplished in yeast by cooperation between SWR1 and NuA4. In agreement with such a model, NuA4 and SWR1 mutants show strong genetic interactions, NuA4 affects histone H2AZ incorporation/acetylation in vivo, and both preset the PHO5 promoter for activation. Interestingly, the expression of a chimeric Eaf1-Swr1 protein recreates a single human-like complex in yeast cells. Our results identified the key central subunit for the structure and functions of the NuA4 histone acetyltransferase complex and functionally linked this activity with the histone variant H2AZ from yeast to human cells.
Figures
References
-
- Altaf, M., N. Saksouk, and J. Cote. 2007. Histone modifications in response to DNA damage. Mutat. Res. 61881-90. - PubMed
-
- Avvakumov, N., and J. Cote. 2007a. Functions of myst family histone acetyltransferases and their link to disease. Subcell. Biochem. 41295-317. - PubMed
-
- Avvakumov, N., and J. Cote. 2007b. The MYST family of histone acetyltransferases and their intimate links to cancer. Oncogene 265395-5407. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
