Epidermal-growth-factor-dependent phosphorylation and ubiquitinylation of MAGE-11 regulates its interaction with the androgen receptor
- PMID: 18212060
- PMCID: PMC2268407
- DOI: 10.1128/MCB.01672-07
Epidermal-growth-factor-dependent phosphorylation and ubiquitinylation of MAGE-11 regulates its interaction with the androgen receptor
Abstract
The androgen receptor (AR) is a ligand-activated transcription factor that interacts with coregulatory proteins during androgen-dependent gene regulation. Melanoma antigen gene protein 11 (MAGE-11) is an AR coregulator that specifically binds the AR NH(2)-terminal FXXLF motif and modulates the AR NH(2)- and carboxyl-terminal N/C interaction to increase AR transcriptional activity. Here we demonstrate that epidermal growth factor (EGF) signaling increases androgen-dependent AR transcriptional activity through the posttranslational modification of MAGE-11. EGF in the presence of dihydrotestosterone stabilizes the AR-MAGE complex through the site-specific phosphorylation of MAGE-11 at Thr-360 and ubiquitinylation at Lys-240 and Lys-245. The time-dependent EGF-induced increase in AR transcriptional activity by MAGE-11 is mediated through AR activation functions 1 and 2 in association with the increased turnover of AR and MAGE-11. The results reveal a dynamic mechanism whereby growth factor signaling increases AR transcriptional activity through the covalent modification of an AR-specific coregulatory protein. Sequence conservation of the MAGE-11 phosphorylation and ubiquitinylation sites throughout the MAGE gene family suggests common regulatory mechanisms for this group of cancer-testis antigens.
Figures


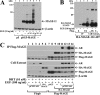

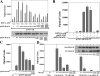





Similar articles
-
Melanoma antigen-A11 (MAGE-A11) enhances transcriptional activity by linking androgen receptor dimers.J Biol Chem. 2013 Jan 18;288(3):1939-52. doi: 10.1074/jbc.M112.428409. Epub 2012 Nov 21. J Biol Chem. 2013. PMID: 23172223 Free PMC article.
-
Transcriptional synergy between melanoma antigen gene protein-A11 (MAGE-11) and p300 in androgen receptor signaling.J Biol Chem. 2010 Jul 9;285(28):21824-36. doi: 10.1074/jbc.M110.120600. Epub 2010 May 6. J Biol Chem. 2010. PMID: 20448036 Free PMC article.
-
Gain in transcriptional activity by primate-specific coevolution of melanoma antigen-A11 and its interaction site in androgen receptor.J Biol Chem. 2011 Aug 26;286(34):29951-63. doi: 10.1074/jbc.M111.244715. Epub 2011 Jul 5. J Biol Chem. 2011. PMID: 21730049 Free PMC article.
-
Structural features discriminate androgen receptor N/C terminal and coactivator interactions.Mol Cell Endocrinol. 2012 Jan 30;348(2):403-10. doi: 10.1016/j.mce.2011.03.026. Epub 2011 Jun 1. Mol Cell Endocrinol. 2012. PMID: 21664945 Free PMC article. Review.
-
Androgen receptor phosphorylation: biological context and functional consequences.Endocr Relat Cancer. 2014 Aug;21(4):T131-45. doi: 10.1530/ERC-13-0472. Epub 2014 Jan 14. Endocr Relat Cancer. 2014. PMID: 24424504 Free PMC article. Review.
Cited by
-
MAGEA11 as a STAD Prognostic Biomarker Associated with Immune Infiltration.Diagnostics (Basel). 2022 Oct 16;12(10):2506. doi: 10.3390/diagnostics12102506. Diagnostics (Basel). 2022. PMID: 36292195 Free PMC article.
-
Site-specific androgen receptor serine phosphorylation linked to epidermal growth factor-dependent growth of castration-recurrent prostate cancer.J Biol Chem. 2008 Jul 25;283(30):20989-1001. doi: 10.1074/jbc.M802392200. Epub 2008 May 29. J Biol Chem. 2008. PMID: 18511414 Free PMC article.
-
Melanoma antigen gene protein-A11 (MAGE-11) F-box links the androgen receptor NH2-terminal transactivation domain to p160 coactivators.J Biol Chem. 2009 Dec 11;284(50):34793-808. doi: 10.1074/jbc.M109.065979. Epub 2009 Oct 14. J Biol Chem. 2009. PMID: 19828458 Free PMC article.
-
Melanoma antigen-11 inhibits the hypoxia-inducible factor prolyl hydroxylase 2 and activates hypoxic response.Cancer Res. 2009 Jan 15;69(2):616-24. doi: 10.1158/0008-5472.CAN-08-0811. Cancer Res. 2009. PMID: 19147576 Free PMC article.
-
Proto-oncogene activity of melanoma antigen-A11 (MAGE-A11) regulates retinoblastoma-related p107 and E2F1 proteins.J Biol Chem. 2013 Aug 23;288(34):24809-24. doi: 10.1074/jbc.M113.468579. Epub 2013 Jul 12. J Biol Chem. 2013. PMID: 23853093 Free PMC article.
References
-
- Bai, S., G. Grossman, L. Yuan, B. A. Lessey, F. S. French, S. L. Young, and E. M. Wilson. 11 December 2007, posting date. Hormone control and expression of androgen receptor coregulator MAGE-11 in human endometrium during the window of receptivity to embryo implantation. Mol. Hum. Reprod. doi:10.1093/molehr/gam080. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
