Generation and characterization of rgs5 mutant mice
- PMID: 18212066
- PMCID: PMC2268431
- DOI: 10.1128/MCB.01252-07
Generation and characterization of rgs5 mutant mice
Abstract
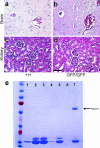
Regulators of G-protein signaling (RGS) are involved in a wide variety of functions, including olfaction, vision, and cell migration. RGS5 has a perivascular expression pattern and was recently identified as a marker for brain pericytes. This suggests a role for RGS5 in vascular development and pericyte biology. We have created a mouse line which lacks the rgs5 gene and replaced it with a green fluorescent protein (GFP) reporter (rgs5(GFP/GFP)). The mice are viable and fertile and display no obvious developmental defects, and the vasculature appears to develop normally with proper pericyte coverage. Also, no differences were observed in the vasculature under pathological conditions, such as tumor growth and oxygen-induced retinopathy. The GFP expression in pericytes of rgs5(GFP) mice allows detection and sorting of these cells, thereby providing a valuable novel tool for pericyte research.
Figures




Similar articles
-
Regulator of G-protein signaling 5 regulates the shift from perivascular to parenchymal pericytes in the chronic phase after stroke.FASEB J. 2019 Aug;33(8):8990-8998. doi: 10.1096/fj.201900153R. Epub 2019 May 2. FASEB J. 2019. PMID: 31039042 Free PMC article.
-
Transcription profiling of platelet-derived growth factor-B-deficient mouse embryos identifies RGS5 as a novel marker for pericytes and vascular smooth muscle cells.Am J Pathol. 2003 Mar;162(3):721-9. doi: 10.1016/S0002-9440(10)63868-0. Am J Pathol. 2003. PMID: 12598306 Free PMC article.
-
RGS5 expression is a quantitative measure of pericyte coverage of blood vessels.Angiogenesis. 2008;11(2):141-51. doi: 10.1007/s10456-007-9085-x. Epub 2007 Nov 25. Angiogenesis. 2008. PMID: 18038251
-
Regulator of G protein signaling 5: a new player in vascular remodeling.Trends Cardiovasc Med. 2009 Jan;19(1):26-30. doi: 10.1016/j.tcm.2009.04.002. Trends Cardiovasc Med. 2009. PMID: 19467451 Review.
-
Review of the pericyte during angiogenesis and its role in cancer and diabetic retinopathy.Toxicol Pathol. 2006;34(6):763-75. doi: 10.1080/01926230600936290. Toxicol Pathol. 2006. PMID: 17162534 Review. No abstract available.
Cited by
-
Regulator of G-protein signaling-5 is a marker of hepatic stellate cells and expression mediates response to liver injury.PLoS One. 2014 Oct 7;9(10):e108505. doi: 10.1371/journal.pone.0108505. eCollection 2014. PLoS One. 2014. PMID: 25290689 Free PMC article.
-
Genetic Analysis of Rare Human Variants of Regulators of G Protein Signaling Proteins and Their Role in Human Physiology and Disease.Pharmacol Rev. 2018 Jul;70(3):446-474. doi: 10.1124/pr.117.015354. Pharmacol Rev. 2018. PMID: 29871944 Free PMC article. Review.
-
Regulator of G-protein signaling 5 (RGS5) protein: a novel marker of cancer vasculature elicited and sustained by the tumor's proangiogenic microenvironment.Cell Mol Life Sci. 2012 Apr;69(7):1167-78. doi: 10.1007/s00018-011-0862-8. Epub 2011 Dec 1. Cell Mol Life Sci. 2012. PMID: 22130514 Free PMC article.
-
Regulator of G-protein signaling 5 regulates the shift from perivascular to parenchymal pericytes in the chronic phase after stroke.FASEB J. 2019 Aug;33(8):8990-8998. doi: 10.1096/fj.201900153R. Epub 2019 May 2. FASEB J. 2019. PMID: 31039042 Free PMC article.
-
Regulator of G Protein Signaling 2: A Versatile Regulator of Vascular Function.Prog Mol Biol Transl Sci. 2015;133:77-92. doi: 10.1016/bs.pmbts.2015.02.001. Epub 2015 Apr 16. Prog Mol Biol Transl Sci. 2015. PMID: 26123303 Free PMC article. Review.
References
-
- Abramsson, A., S. Kurup, M. Busse, S. Yamada, P. Lindblom, E. Schallmeiner, D. Stenzel, D. Sauvaget, J. Ledin, M. Ringvall, U. Landegren, L. Kjellen, G. Bondjers, J. P. Li, U. Lindahl, D. Spillmann, C. Betsholtz, and H. Gerhardt. 2007. Defective N-sulfation of heparan sulfate proteoglycans limits PDGF-BB binding and pericyte recruitment in vascular development. Genes Dev. 21316-331. - PMC - PubMed
-
- Berger, M., G. Bergers, B. Arnold, G. J. Hammerling, and R. Ganss. 2005. Regulator of G-protein signaling-5 induction in pericytes coincides with active vessel remodeling during neovascularization. Blood 1051094-1101. - PubMed
-
- Berthebaud, M., C. Riviere, P. Jarrier, A. Foudi, Y. Zhang, D. Compagno, A. Galy, W. Vainchenker, and F. Louache. 2005. RGS16 is a negative regulator of SDF-1-CXCR4 signaling in megakaryocytes. Blood 1062962-2968. - PubMed
-
- Bodenstein, J., R. K. Sunahara, and R. R. Neubig. 2007. N-terminal residues control proteasomal degradation of RGS2, RGS4, and RGS5 in human embryonic kidney 293 cells. Mol. Pharmacol. 711040-1050. - PubMed
-
- Bondjers, C., L. He, M. Takemoto, J. Norlin, N. Asker, M. Hellstrom, P. Lindahl, and C. Betsholtz. 2006. Microarray analysis of blood microvessels from PDGF-B and PDGF-Rbeta mutant mice identifies novel markers for brain pericytes. FASEB J. 201703-1705. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
