In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells
- PMID: 18212077
- PMCID: PMC2292855
- DOI: 10.1128/IAI.01373-07
In vitro analysis of tobramycin-treated Pseudomonas aeruginosa biofilms on cystic fibrosis-derived airway epithelial cells
Abstract
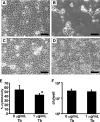
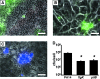
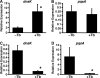
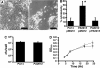
P. aeruginosa forms biofilms in the lungs of individuals with cystic fibrosis (CF); however, there have been no effective model systems for studying biofilm formation in the CF lung. We have developed a tissue culture system for growth of P. aeruginosa biofilms on CF-derived human airway cells that promotes the formation of highly antibiotic-resistant microcolonies, which produce an extracellular polysaccharide matrix and require the known abiotic biofilm formation genes flgK and pilB. Treatment of P. aeruginosa biofilms with tobramycin reduced the virulence of the biofilms both by reducing bacterial numbers and by altering virulence gene expression. We performed microarray analysis of these biofilms on epithelial cells after treatment with tobramycin, and we compared these results with gene expression of (i) tobramycin-treated planktonic P. aeruginosa and (ii) tobramycin-treated P. aeruginosa biofilms on an abiotic surface. Despite the conservation in functions required to form a biofilm, our results show that the responses to tobramycin treatment of biofilms grown on biotic versus abiotic surfaces are different, as exemplified by downregulation of genes involved in Pseudomonas quinolone signal biosynthesis specifically in epithelial cell-grown biofilms versus plastic-grown biofilms. We also identified the gene PA0913, which is upregulated by tobramycin specifically in biofilms grown on CF airway cells and codes for a probable magnesium transporter, MgtE. Mutation of the PA0913 gene increased the bacterial virulence of biofilms on the epithelial cells, consistent with a role for the gene in the suppression of bacterial virulence. Taken together, our data show that analysis of biofilms on airway cells provides new insights into the interaction of these microbial communities with the host.
Figures


References
-
- Boddicker, J. D., N. A. Ledeboer, J. Jagnow, B. D. Jones, and S. Clegg. 2002. Differential binding to and biofilm formation on, HEp-2 cells by Salmonella enterica serovar Typhimurium is dependent upon allelic variation in the fimH gene of the fim gene cluster. Mol. Microbiol. 451255-1265. - PubMed
-
- Boucher, R. C. 2004. New concepts of the pathogenesis of cystic fibrosis lung disease. Eur. Respir. J. 23146-158. - PubMed
-
- Bruscia, E., F. Sangiuolo, P. Sinibaldi, K. K. Goncz, G. Novelli, and D. C. Gruenert. 2002. Isolation of CF cell lines corrected at DeltaF508-CFTR locus by SFHR-mediated targeting. Gene Ther. 9683-685. - PubMed
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

