Genomic analysis reveals Mycoplasma pneumoniae repetitive element 1-mediated recombination in a clinical isolate
- PMID: 18212079
- PMCID: PMC2292888
- DOI: 10.1128/IAI.01621-07
Genomic analysis reveals Mycoplasma pneumoniae repetitive element 1-mediated recombination in a clinical isolate
Abstract
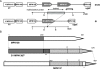
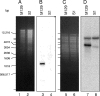
Mycoplasmas are cell wall-less bacteria that evolved by drastic reduction of the genome size. Complete genome analysis of Mycoplasma pneumoniae revealed the presence of numerous copies of four distinct large M. pneumoniae repetitive elements (RepMPs). One copy each of RepMP2/3, RepMP4, and RepMP5 are localized within the P1 operon (MPN140 to MPN142 loci), and their involvement in sequence variation in adhesin P1 and adherence-related protein B/C has been documented. Here we analyzed a clinical strain of M. pneumoniae designated S1 isolated from a 1993 outbreak of respiratory infections in San Antonio, TX. Based on the type of RepMPs within the P1 operon, we classified clinical isolate S1 as type 2 with unique minor sequence variations. Hybridization with oligonucleotide arrays revealed sequence divergence in two previously unsuspected hypothetical genes (MPN137 and MPN138 loci). Closer inspection of this region revealed that the MPN137 and MPN138 loci harbored previously unrecognized unique RepMP1 sequences found only in M. pneumoniae. PCR and sequence analyses revealed a recombination event involving three RepMP1-containing genes that resulted in fusion of MPN137 and MPN138 reading frames and loss of all but a short fragment of another RepMP1-containing locus, MPN130. The multiple copies of unique RepMP1 elements spread throughout the chromosome could allow vast numbers of sequence variations in clinical strains. Comparisons of amino acid sequences showed the presence of leucine zipper motifs in MPN130 and MPN138 proteins in reference strain M129 and the absence of these motifs in the fused protein of S1. The presence of tandem leucine and other repeats points to possible regulatory functions of proteins encoded by RepMP1-containing genes.
Figures



References
-
- Barr, F. G., N. Galili, J. Holick, J. A. Biegel, G. Rovera, and B. S. Emanuel. 1993. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 3113-117. - PubMed
-
- Buckland, R., and F. Wild. 1989. Leucine zipper motif extends. Nature 338547. - PubMed
-
- Cousin, A., B. de Barbeyrac, A. Charron, H. Renadin, and C. Bebear. 1994. Analysis of RFLPs of amplified cytadhesin P1 gene for epidemiological study of Mycoplasma pneumoniae. IOM Lett. 3494-495.
-
- Cousin-Allery, A., A. Charron, B. de Barbeyrac, G. Fremy, J. Skov Jensen, H. Renaudin, and C. Bebear. 2000. Molecular typing of Mycoplasma pneumoniae strains by PCR-based methods and pulsed-field gel electrophoresis. Application to French and Danish isolates. Epidemiol. Infect. 124103-111. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

