Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus
- PMID: 18212094
- PMCID: PMC2292509
- DOI: 10.1128/AAC.01164-07
Spontaneous deletion of the methicillin resistance determinant, mecA, partially compensates for the fitness cost associated with high-level vancomycin resistance in Staphylococcus aureus
Abstract
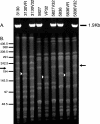
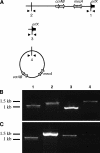
Treatment of infections caused by Staphylococcus aureus is often confounded by the bacterium's ability to develop resistance to chemotherapeutic agents. Methicillin-resistant S. aureus (MRSA) arises through the acquisition of staphylococcal chromosomal cassette mec (SCCmec), a genomic island containing the methicillin resistance determinant, mecA. In contrast, resistance to vancomycin can result from exposure to the drug, a mechanism that is not dependent upon a gene acquisition event. Here we describe three MRSA strains that became resistant to vancomycin during passage in the presence of increasing concentrations of the drug. In each case two derivative strains were isolated, one that had lost mecA and one that retained mecA during passage. Strain 5836VR lost mecA by the site-specific chromosomal excision of SCCmec, while the other two strains (strains 3130VR and VP32) deleted portions of their SCCmec elements in a manner that appeared to involve IS431. Conversion to vancomycin resistance caused a decrease in the growth rate that was partially compensated for by the deletion of mecA. In mixed-culture competition experiments, vancomycin-resistant strains that lacked mecA readily outcompeted their mecA-containing counterparts, suggesting that the loss of mecA during conversion to vancomycin resistance was advantageous to the organism.
Figures





References
-
- Adhikari, R. P., G. C. Scales, K. Kobayashi, J. M. Smith, B. Berger-Bachi, and G. M. Cook. 2004. Vancomycin-induced deletion of the methicillin resistance gene mecA in Staphylococcus aureus. J. Antimicrob. Chemother. 54:360-363. - PubMed
-
- Berger-Bachi, B. 1994. Expression of resistance to methicillin. Trends Microbiol. 2:389-393. - PubMed
-
- Berger-Bachi, B. 1997. Resistance not mediated by beta-lactamase (methicillin resistance), p. 158-174. In K. B. Crossley and G. L. Archer (ed.), The staphylococci in human disease. Churchill Livingstone Inc., New York, NY.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

