The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples
- PMID: 18213365
- PMCID: PMC2186209
- DOI: 10.1371/journal.pone.0001456
The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples
Abstract
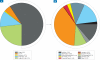
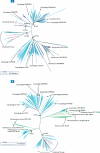
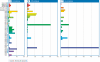
Viruses are the most abundant biological entities on our planet. Interactions between viruses and their hosts impact several important biological processes in the world's oceans such as horizontal gene transfer, microbial diversity and biogeochemical cycling. Interrogation of microbial metagenomic sequence data collected as part of the Sorcerer II Global Ocean Expedition (GOS) revealed a high abundance of viral sequences, representing approximately 3% of the total predicted proteins. Cluster analyses of the viral sequences revealed hundreds to thousands of viral genes encoding various metabolic and cellular functions. Quantitative analyses of viral genes of host origin performed on the viral fraction of aquatic samples confirmed the viral nature of these sequences and suggested that significant portions of aquatic viral communities behave as reservoirs of such genetic material. Distributional and phylogenetic analyses of these host-derived viral sequences also suggested that viral acquisition of environmentally relevant genes of host origin is a more abundant and widespread phenomenon than previously appreciated. The predominant viral sequences identified within microbial fractions originated from tailed bacteriophages and exhibited varying global distributions according to viral family. Recruitment of GOS viral sequence fragments against 27 complete aquatic viral genomes revealed that only one reference bacteriophage genome was highly abundant and was closely related, but not identical, to the cyanomyovirus P-SSM4. The co-distribution across all sampling sites of P-SSM4-like sequences with the dominant ecotype of its host, Prochlorococcus supports the classification of the viral sequences as P-SSM4-like and suggests that this virus may influence the abundance, distribution and diversity of one of the most dominant components of picophytoplankton in oligotrophic oceans. In summary, the abundance and broad geographical distribution of viral sequences within microbial fractions, the prevalence of genes among viral sequences that encode microbial physiological function and their distinct phylogenetic distribution lend strong support to the notion that viral-mediated gene acquisition is a common and ongoing mechanism for generating microbial diversity in the marine environment.
Conflict of interest statement
Figures


References
-
- Bergh O, Borsheim KY, Bratbak G, Heldal M. High Abundance of Viruses Found in Aquatic Environments. Nature. 1989;340:467–468. - PubMed
-
- Fuhrman JA. Marine viruses and their biogeochemical and ecological effects. Nature. 1999;399:541–548. - PubMed
-
- Shibata A, Kogure K, Koike I, Ohwada K. Formation of submicron colloidal particles from marine bacteria by viral infection. Marine Ecology-Progress Series. 1997;155:303–307.
-
- Fuhrman JA, Wilcox RM, Noble RT, Law NC. Viruses in marine food webs. In: Guerrero R, Pedros-Alio C, editors. Barcelona, Spain: Trends Microbial Ecol; 1993. pp. 295–298.
-
- Gobler CJ, Hutchins DA, Fisher NS, Cosper EM, Sanudo-Wilhelmy SA. Release and bioavailability of C, N, P, Se, and Fe following viral lysis of a marine chrysophyte. Limnology and Oceanography. 1997;42:1492–1504.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

