The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron
- PMID: 18214421
- PMCID: PMC2587239
- DOI: 10.1007/s00122-007-0706-y
The complete nucleotide sequence of the cassava (Manihot esculenta) chloroplast genome and the evolution of atpF in Malpighiales: RNA editing and multiple losses of a group II intron
Abstract
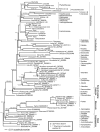
The complete sequence of the chloroplast genome of cassava (Manihot esculenta, Euphorbiaceae) has been determined. The genome is 161,453 bp in length and includes a pair of inverted repeats (IR) of 26,954 bp. The genome includes 128 genes; 96 are single copy and 16 are duplicated in the IR. There are four rRNA genes and 30 distinct tRNAs, seven of which are duplicated in the IR. The infA gene is absent; expansion of IRb has duplicated 62 amino acids at the 3' end of rps19 and a number of coding regions have large insertions or deletions, including insertions within the 23S rRNA gene. There are 17 intron-containing genes in cassava, 15 of which have a single intron while two (clpP, ycf3) have two introns. The usually conserved atpF group II intron is absent and this is the first report of its loss from land plant chloroplast genomes. The phylogenetic distribution of the atpF intron loss was determined by a PCR survey of 251 taxa representing 34 families of Malpighiales and 16 taxa from closely related rosids. The atpF intron is not only missing in cassava but also from closely related Euphorbiaceae and other Malpighiales, suggesting that there have been at least seven independent losses. In cassava and all other sequenced Malphigiales, atpF gene sequences showed a strong association between C-to-T substitutions at nucleotide position 92 and the loss of the intron, suggesting that recombination between an edited mRNA and the atpF gene may be a possible mechanism for the intron loss.
Figures



References
-
- APG . An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot J Linn Soc. 2003;141:399–436.
-
- Barkan A. Intron splicing in plant organelles. In: Daniell H, Chase CD, editors. Molecular biology and biotechnology of plant organelles: chloroplast and mitochondria. Springer; Netherlands: 2004. pp. 295–322.
-
- Berry PE, Hipp AL, Wurdack KJ, van Ee B, Riina R. Molecular phylogenetics of the giant genus Croton and tribe Crotoneae (Euphorbiaceae sensu stricto) using ITS and trnL-trnF DNA sequence data. Am J Bot. 2005;92:1520–1534. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

