I-TASSER server for protein 3D structure prediction
- PMID: 18215316
- PMCID: PMC2245901
- DOI: 10.1186/1471-2105-9-40
I-TASSER server for protein 3D structure prediction
Abstract
Background: Prediction of 3-dimensional protein structures from amino acid sequences represents one of the most important problems in computational structural biology. The community-wide Critical Assessment of Structure Prediction (CASP) experiments have been designed to obtain an objective assessment of the state-of-the-art of the field, where I-TASSER was ranked as the best method in the server section of the recent 7th CASP experiment. Our laboratory has since then received numerous requests about the public availability of the I-TASSER algorithm and the usage of the I-TASSER predictions.
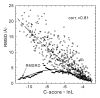
Results: An on-line version of I-TASSER is developed at the KU Center for Bioinformatics which has generated protein structure predictions for thousands of modeling requests from more than 35 countries. A scoring function (C-score) based on the relative clustering structural density and the consensus significance score of multiple threading templates is introduced to estimate the accuracy of the I-TASSER predictions. A large-scale benchmark test demonstrates a strong correlation between the C-score and the TM-score (a structural similarity measurement with values in [0, 1]) of the first models with a correlation coefficient of 0.91. Using a C-score cutoff > -1.5 for the models of correct topology, both false positive and false negative rates are below 0.1. Combining C-score and protein length, the accuracy of the I-TASSER models can be predicted with an average error of 0.08 for TM-score and 2 A for RMSD.
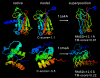
Conclusion: The I-TASSER server has been developed to generate automated full-length 3D protein structural predictions where the benchmarked scoring system helps users to obtain quantitative assessments of the I-TASSER models. The output of the I-TASSER server for each query includes up to five full-length models, the confidence score, the estimated TM-score and RMSD, and the standard deviation of the estimations. The I-TASSER server is freely available to the academic community at http://zhang.bioinformatics.ku.edu/I-TASSER.
Figures


Similar articles
-
Ab initio modeling of small proteins by iterative TASSER simulations.BMC Biol. 2007 May 8;5:17. doi: 10.1186/1741-7007-5-17. BMC Biol. 2007. PMID: 17488521 Free PMC article.
-
LOMETS: a local meta-threading-server for protein structure prediction.Nucleic Acids Res. 2007;35(10):3375-82. doi: 10.1093/nar/gkm251. Epub 2007 May 3. Nucleic Acids Res. 2007. PMID: 17478507 Free PMC article.
-
Template-based protein structure prediction in CASP11 and retrospect of I-TASSER in the last decade.Proteins. 2016 Sep;84 Suppl 1(Suppl 1):233-46. doi: 10.1002/prot.24918. Epub 2015 Sep 18. Proteins. 2016. PMID: 26343917 Free PMC article.
-
AI-Driven Deep Learning Techniques in Protein Structure Prediction.Int J Mol Sci. 2024 Aug 1;25(15):8426. doi: 10.3390/ijms25158426. Int J Mol Sci. 2024. PMID: 39125995 Free PMC article. Review.
-
Critical evaluation of in silico methods for prediction of coiled-coil domains in proteins.Brief Bioinform. 2016 Mar;17(2):270-82. doi: 10.1093/bib/bbv047. Epub 2015 Jul 15. Brief Bioinform. 2016. PMID: 26177815 Free PMC article. Review.
Cited by
-
A cucumber mosaic virus based expression system for the production of porcine circovirus specific vaccines.PLoS One. 2012;7(12):e52688. doi: 10.1371/journal.pone.0052688. Epub 2012 Dec 20. PLoS One. 2012. PMID: 23285149 Free PMC article.
-
Sequence conservation, phylogenetic relationships, and expression profiles of nondigestive serine proteases and serine protease homologs in Manduca sexta.Insect Biochem Mol Biol. 2015 Jul;62:51-63. doi: 10.1016/j.ibmb.2014.10.006. Epub 2014 Dec 19. Insect Biochem Mol Biol. 2015. PMID: 25530503 Free PMC article.
-
Structural insight into HIV-1 capsid recognition by rhesus TRIM5α.Proc Natl Acad Sci U S A. 2012 Nov 6;109(45):18372-7. doi: 10.1073/pnas.1210903109. Epub 2012 Oct 22. Proc Natl Acad Sci U S A. 2012. PMID: 23091002 Free PMC article.
-
Preclinical Validation of the Heparin-Reactive Peptide p5+14 as a Molecular Imaging Agent for Visceral Amyloidosis.Molecules. 2015 Apr 27;20(5):7657-82. doi: 10.3390/molecules20057657. Molecules. 2015. PMID: 25923515 Free PMC article.
-
Transient Worsening of Photosensitivity due to Cholelithiasis in a Variegate Porphyria Patient.Intern Med. 2016;55(20):2965-2969. doi: 10.2169/internalmedicine.55.7108. Epub 2016 Oct 15. Intern Med. 2016. PMID: 27746433 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

