Electron cryomicroscopy reveals different F1+F2 protein States in intact parainfluenza virions
- PMID: 18216117
- PMCID: PMC2268498
- DOI: 10.1128/JVI.02154-07
Electron cryomicroscopy reveals different F1+F2 protein States in intact parainfluenza virions
Abstract
Electron cryomicrographs of intact parainfluenza virus 5 (PIV5) virions revealed two different surface structures, namely, a continuous layer and distinct individual spikes. The structure of these spikes reconstructed from intact virions was compared with known F ectodomain structures and was found to be different from the prefusion PIV5 F0 structure but, surprisingly, very similar to the human PIV3 F postfusion structure. Hence, we conclude that the individual F1+F2 spikes in intact PIV5 virions also correspond to the postfusion state. Since the observed fusion activity of PIV5 virions has to be associated with prefusion F1+F2 proteins, they have necessarily to be localized in the continuous surface structure. The data therefore strongly suggest that the prefusion state of the F1+F2 protein requires stabilization, most probably by the association with hemagglutinin-neuraminidase. The conversion of F1+F2 proteins from the prefusion toward the postfusion state while embedded in the virus membrane is topologically difficult to comprehend on the basis of established models and demands reconsideration of our current understanding.
Figures


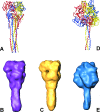
References
-
- Baker, K. A., R. E. Dutch, R. A. Lamb, and T. S. Jardetzky. 1999. Structural basis for paramyxovirus-mediated membrane fusion. Mol. Cell 3309-319. - PubMed
-
- Blumenthal, R., A. Bali-Puri, A. Walter, D. Covell, and O. Eidelman. 1987. pH-dependent fusion of vesicular stomatitis virus with Vero cells. Measurement by dequenching of octadecyl rhodamine fluorescence. J. Biol. Chem. 26213614-13619. - PubMed
-
- Böttcher, C., K. Ludwig, A. Herrmann, M. van Heel, and H. Stark. 1999. Structure of influenza hemagglutinin at neutral and at fusogenic pH by electron cryo-microscopy. FEBS Lett. 463255-259. - PubMed
-
- Chan, D. C., and P. S. Kim. 1998. HIV entry and its inhibition. Cell 93681-684. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

