Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel
- PMID: 18216238
- PMCID: PMC2234175
- DOI: 10.1073/pnas.0711533105
Structure of the transmembrane regions of a bacterial cyclic nucleotide-regulated channel
Abstract
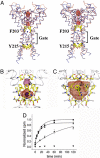
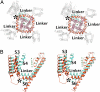
The six-transmembrane helix (6 TM) tetrameric cation channels form the largest ion channel family, some members of which are voltage-gated and others are not. There are no reported channel structures to match the wealth of functional data on the non-voltage-gated members. We determined the structure of the transmembrane regions of the bacterial cyclic nucleotide-regulated channel MlotiK1, a non-voltage-gated 6 TM channel. The structure showed how the S1-S4 domain and its associated linker can serve as a clamp to constrain the gate of the pore and possibly function in concert with ligand-binding domains to regulate the opening of the pore. The structure also led us to hypothesize a new mechanism by which motions of the S6 inner helices can gate the ion conduction pathway at a position along the pore closer to the selectivity filter than the canonical helix bundle crossing.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

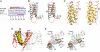
References
-
- Hille B. Ion Channels of Excitable Membranes. Sunderland, MA: Sinauer; 2001.
-
- Jan LY, Jan YN. Tracing the roots of ion channels. Cell. 1992;69:715–718. - PubMed
-
- Sigworth FJ. Voltage gating of ion channels. Q Rev Biophys. 1994;27:1–40. - PubMed
-
- Aggarwal SK, MacKinnon R. Contribution of the S4 segment to gating charge in the Shaker K+ channel. Neuron. 1996;16:1169–1177. - PubMed
-
- Seoh SA, Sigg D, Papazian DM, Bezanilla F. Voltage-sensing residues in the S2 and S4 segments of the Shaker K+ channel. Neuron. 1996;16:1159–1167. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

