Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification
- PMID: 18216255
- PMCID: PMC2234137
- DOI: 10.1073/pnas.0710217105
Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification
Abstract
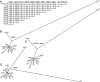
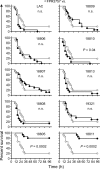
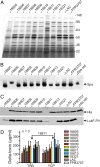
Emerging and re-emerging infectious diseases, especially those caused by drug-resistant bacteria, are a major problem worldwide. Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) appeared rapidly and unexpectedly in the United States, resulting in an epidemic caused primarily by isolates classified as USA300. The evolutionary and molecular underpinnings of this epidemic are poorly understood. Specifically, it is unclear whether there has been clonal emergence of USA300 isolates or evolutionary convergence toward a hypervirulent phenotype resulting in the independent appearance of similar organisms. To definitively resolve this issue and understand the phylogeny of USA300 isolates, we used comparative whole-genome sequencing to analyze 10 USA300 patient isolates from eight states in diverse geographic regions of the United States and multiple types of human infection. Eight of 10 isolates analyzed had very few single nucleotide polymorphisms (SNPs) and thus were closely related, indicating recent diversification rather than convergence. Unexpectedly, 2 of the clonal isolates had significantly reduced mortality in a mouse sepsis model compared with the reference isolate (P = 0.0002), providing strong support to the idea that minimal genetic change in the bacterial genome can have profound effects on virulence. Taken together, our results demonstrate that there has been recent clonal expansion and diversification of a subset of isolates classified as USA300. The findings add an evolutionary dimension to the epidemiology and emergence of USA300 and suggest a similar mechanism for the pandemic occurrence and spread of penicillin-resistant S. aureus (known as phage-type 80/81 S. aureus) in the 1950s.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

