UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence
- PMID: 18216277
- PMCID: PMC2291399
- DOI: 10.1091/mbc.e07-11-1110
UME6, a novel filament-specific regulator of Candida albicans hyphal extension and virulence
Abstract
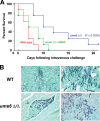
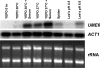
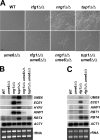
The specific ability of the major human fungal pathogen Candida albicans, as well as many other pathogenic fungi, to extend initial short filaments (germ tubes) into elongated hyphal filaments is important for a variety of virulence-related processes. However, the molecular mechanisms that control hyphal extension have remained poorly understood for many years. We report the identification of a novel C. albicans transcriptional regulator, UME6, which is induced in response to multiple host environmental cues and is specifically important for hyphal extension. Although capable of forming germ tubes, the ume6Delta/ume6Delta mutant exhibits a clear defect in hyphal extension both in vitro and during infection in vivo and is attenuated for virulence in a mouse model of systemic candidiasis. We also show that UME6 is an important downstream component of both the RFG1-TUP1 and NRG1-TUP1 filamentous growth regulatory pathways, and we provide evidence to suggest that Nrg1 and Ume6 function together by a negative feedback loop to control the level and duration of filament-specific gene expression in response to inducing conditions. Our results suggest that hyphal extension is controlled by a specific transcriptional regulatory mechanism and is correlated with the maintenance of high-level expression of genes in the C. albicans filamentous growth program.
Figures


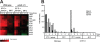


References
-
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., editors. New York: Greene Publishing Associates and Wiley-Interscience; 1992. Current Protocols in Molecular Biology.
-
- Bartie K. L., Williams D. W., Wilson M. J., Potts A. J., Lewis M. A. Differential invasion of Candida albicans isolates in an in vitro model of oral candidosis. Oral Microbiol. Immunol. 2004;19:293–296. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

