Comparison of Mascot and X!Tandem performance for low and high accuracy mass spectrometry and the development of an adjusted Mascot threshold
- PMID: 18216375
- PMCID: PMC2656932
- DOI: 10.1074/mcp.M700293-MCP200
Comparison of Mascot and X!Tandem performance for low and high accuracy mass spectrometry and the development of an adjusted Mascot threshold
Abstract
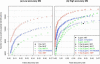
It is a major challenge to develop effective sequence database search algorithms to translate molecular weight and fragment mass information obtained from tandem mass spectrometry into high quality peptide and protein assignments. We investigated the peptide identification performance of Mascot and X!Tandem for mass tolerance settings common for low and high accuracy mass spectrometry. We demonstrated that sensitivity and specificity of peptide identification can vary substantially for different mass tolerance settings, but this effect was more significant for Mascot. We present an adjusted Mascot threshold, which allows the user to freely select the best trade-off between sensitivity and specificity. The adjusted Mascot threshold was compared with the default Mascot and X!Tandem scoring thresholds and shown to be more sensitive at the same false discovery rates for both low and high accuracy mass spectrometry data.
Figures



References
-
- Rudnick P, Wang Y, Evans E, Lee C, Balgley B. Large scale analysis of MASCOT results using a Mass Accuracy-based THreshold (MATH) effectively improves data interpretation. J. Proteome Res. 2005;4:1353–1360. - PubMed
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. - PubMed
-
- Foster L, de Hoog C, Zhang Y, Zhang Y, Xie X, Mootha V, Mann M. A mammalian organelle map by protein correlation profiling. Cell. 2006;125:187–199. - PubMed
-
- Washburn M, Wolters D, Yates JR. Large-scale analysis of the yeast proteome by multidimensional protein identification technology. Nat. Biotechnol. 2001;19:242–247. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

