Nonribosomal peptide synthetases involved in the production of medically relevant natural products
- PMID: 18217713
- PMCID: PMC3131160
- DOI: 10.1021/mp700137g
Nonribosomal peptide synthetases involved in the production of medically relevant natural products
Abstract
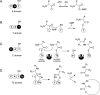
Natural products biosynthesized wholly or in part by nonribosomal peptide synthetases (NRPSs) are some of the most important drugs currently used clinically for the treatment of a variety of diseases. Since the initial research into NRPSs in the early 1960s, we have gained considerable insights into the mechanism by which these enzymes assemble these natural products. This review will present a brief history of how the basic mechanistic steps of NRPSs were initially deciphered and how this information has led us to understand how nature modified these systems to generate the enormous structural diversity seen in nonribosomal peptides. This review will also briefly discuss how drug development and discovery are being influenced by what we have learned from nature about nonribosomal peptide biosynthesis.
Figures
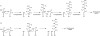
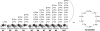
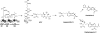
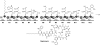
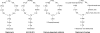
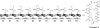

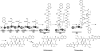
Similar articles
-
Nonribosomal Peptide Synthesis-Principles and Prospects.Angew Chem Int Ed Engl. 2017 Mar 27;56(14):3770-3821. doi: 10.1002/anie.201609079. Epub 2017 Mar 21. Angew Chem Int Ed Engl. 2017. PMID: 28323366 Review.
-
Nonribosomal peptide synthetases: structures and dynamics.Curr Opin Struct Biol. 2010 Apr;20(2):234-40. doi: 10.1016/j.sbi.2010.01.009. Epub 2010 Feb 10. Curr Opin Struct Biol. 2010. PMID: 20153164 Review.
-
Recent progress in the reprogramming of nonribosomal peptide synthetases.J Pept Sci. 2024 Mar;30(3):e3545. doi: 10.1002/psc.3545. Epub 2023 Sep 18. J Pept Sci. 2024. PMID: 37721208 Review.
-
Nonribosomal peptide synthetases and their biotechnological potential in Penicillium rubens.J Ind Microbiol Biotechnol. 2021 Aug 24;48(7-8):kuab045. doi: 10.1093/jimb/kuab045. J Ind Microbiol Biotechnol. 2021. PMID: 34279620 Free PMC article. Review.
-
[Advances in the study of the mechanism and application of nonribosomal peptide synthetases].Wei Sheng Wu Xue Bao. 2007 Aug;47(4):734-7. Wei Sheng Wu Xue Bao. 2007. PMID: 17944384 Review. Chinese.
Cited by
-
Innovative Biomedical and Technological Strategies for the Control of Bacterial Growth and Infections.Biomedicines. 2024 Jan 13;12(1):176. doi: 10.3390/biomedicines12010176. Biomedicines. 2024. PMID: 38255281 Free PMC article. Review.
-
Computational Insights into Amide Bond Formation Catalyzed by the Condensation Domain of Nonribosomal Peptide Synthetases.ACS Omega. 2024 Jun 22;9(26):28556-28563. doi: 10.1021/acsomega.4c02531. eCollection 2024 Jul 2. ACS Omega. 2024. PMID: 38973878 Free PMC article.
-
Feature sequence-based genome mining uncovers the hidden diversity of bacterial siderophore pathways.Elife. 2024 Oct 1;13:RP96719. doi: 10.7554/eLife.96719. Elife. 2024. PMID: 39352117 Free PMC article.
-
Loci Encoding Compounds Potentially Active against Drug-Resistant Pathogens amidst a Decreasing Pool of Novel Antibiotics.Appl Environ Microbiol. 2019 Nov 14;85(23):e01438-19. doi: 10.1128/AEM.01438-19. Print 2019 Dec 1. Appl Environ Microbiol. 2019. PMID: 31540982 Free PMC article.
-
Investigating and Optimizing the Lysate-Based Expression of Nonribosomal Peptide Synthetases Using a Reporter System.ACS Synth Biol. 2023 May 19;12(5):1447-1460. doi: 10.1021/acssynbio.2c00658. Epub 2023 Apr 11. ACS Synth Biol. 2023. PMID: 37039644 Free PMC article.
References
-
- Borowska ZK, Tatum EL. Biosynthesis of edeine by Bacillus brevis Vm4 in vivo and in vitro. Biochim. Biophys. Acta. 1966;114(1):206–9. - PubMed
-
- Yukioka M, Tsukamoto Y, Saito Y, Tsuji T, Otani S, Otani S. Biosynthesis of gramicidin S by a cell-free system of Bacillus brevis. Biochem. Biophys. Res. Commun. 1965;19:204–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

