Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling
- PMID: 18218067
- PMCID: PMC2395256
- DOI: 10.1186/gb-2008-9-1-r17
Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling
Abstract
Background: Dendritic cells (DCs) are a complex group of cells that play a critical role in vertebrate immunity. Lymph-node resident DCs (LN-DCs) are subdivided into conventional DC (cDC) subsets (CD11b and CD8alpha in mouse; BDCA1 and BDCA3 in human) and plasmacytoid DCs (pDCs). It is currently unclear if these various DC populations belong to a unique hematopoietic lineage and if the subsets identified in the mouse and human systems are evolutionary homologs. To gain novel insights into these questions, we sought conserved genetic signatures for LN-DCs and in vitro derived granulocyte-macrophage colony stimulating factor (GM-CSF) DCs through the analysis of a compendium of genome-wide expression profiles of mouse or human leukocytes.
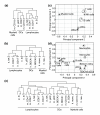
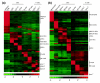
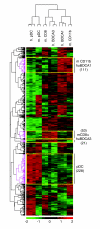
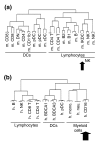
Results: We show through clustering analysis that all LN-DC subsets form a distinct branch within the leukocyte family tree, and reveal a transcriptomal signature evolutionarily conserved in all LN-DC subsets. Moreover, we identify a large gene expression program shared between mouse and human pDCs, and smaller conserved profiles shared between mouse and human LN-cDC subsets. Importantly, most of these genes have not been previously associated with DC function and many have unknown functions. Finally, we use compendium analysis to re-evaluate the classification of interferon-producing killer DCs, lin-CD16+HLA-DR+ cells and in vitro derived GM-CSF DCs, and show that these cells are more closely linked to natural killer and myeloid cells, respectively.
Conclusion: Our study provides a unique database resource for future investigation of the evolutionarily conserved molecular pathways governing the ontogeny and functions of leukocyte subsets, especially DCs.
Figures
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

