Universal primers that amplify RNA from all three flavivirus subgroups
- PMID: 18218114
- PMCID: PMC2263041
- DOI: 10.1186/1743-422X-5-16
Universal primers that amplify RNA from all three flavivirus subgroups
Abstract
Background: Species within the Flavivirus genus pose public health problems around the world. Increasing cases of Dengue and Japanese encephalitis virus in Asia, frequent outbreaks of Yellow fever virus in Africa and South America, and the ongoing spread of West Nile virus throughout the Americas, show the geographical burden of flavivirus diseases. Flavivirus infections are often indistinct from and confused with other febrile illnesses. Here we review the specificity of published primers, and describe a new universal primer pair that can detect a wide range of flaviviruses, including viruses from each of the recognised subgroups.
Results: Bioinformatic analysis of 257 published full-length Flavivirus genomes revealed conserved regions not previously targeted by primers. Two degenerate primers, Flav100F and Flav200R were designed from these regions and used to generate an 800 base pair cDNA product. The region amplified encoded part of the methyltransferase and most of the RNA-dependent-RNA-polymerase (NS5) coding sequence. One-step RT-PCR testing was successful using standard conditions with RNA from over 60 different flavivirus strains representing about 50 species. The cDNA from each virus isolate was sequenced then used in phylogenetic analyses and database searches to confirm the identity of the template RNA.
Conclusion: Comprehensive testing has revealed the broad specificity of these primers. We briefly discuss the advantages and uses of these universal primers.
Figures
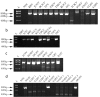

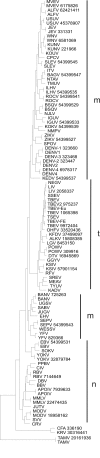
References
-
- Deubel V, Laille M, Hugnot JP, Chungue E, Guesdon JL, Drouet MT, Bassot S, Chevrier D. Identification of dengue sequences by genomic amplification: rapid diagnosis of dengue virus serotypes in peripheral blood. Journal of Virological Methods. 1990;30:41–54. doi: 10.1016/0166-0934(90)90042-E. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

