Structure of a small-molecule inhibitor complexed with GlmU from Haemophilus influenzae reveals an allosteric binding site
- PMID: 18218712
- PMCID: PMC2248321
- DOI: 10.1110/ps.073271408
Structure of a small-molecule inhibitor complexed with GlmU from Haemophilus influenzae reveals an allosteric binding site
Abstract
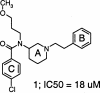
N-Acetylglucosamine-1-phosphate uridyltransferase (GlmU) is an essential enzyme in aminosugars metabolism and an attractive target for antibiotic drug discovery. GlmU catalyzes the formation of uridine-diphospho-N-acetylglucosamine (UDP-GlcNAc), an important precursor in the peptidoglycan and lipopolisaccharide biosynthesis in both Gram-negative and Gram-positive bacteria. Here we disclose a 1.9 A resolution crystal structure of a synthetic small-molecule inhibitor of GlmU from Haemophilus influenzae (hiGlmU). The compound was identified through a high-throughput screening (HTS) configured to detect inhibitors that target the uridyltransferase active site of hiGlmU. The original HTS hit exhibited a modest micromolar potency (IC(50) approximately 18 microM in a racemic mixture) against hiGlmU and no activity against Staphylococcus aureus GlmU (saGlmU). The determined crystal structure indicated that the inhibitor occupies an allosteric site adjacent to the GlcNAc-1-P substrate-binding region. Analysis of the mechanistic model of the uridyltransferase reaction suggests that the binding of this allosteric inhibitor prevents structural rearrangements that are required for the enzymatic reaction, thus providing a basis for structure-guided design of a new class of mechanism-based inhibitors of GlmU.
Figures


Similar articles
-
Characterization of substrate binding and catalysis in the potential antibacterial target N-acetylglucosamine-1-phosphate uridyltransferase (GlmU).Protein Sci. 2007 Dec;16(12):2657-66. doi: 10.1110/ps.073135107. Protein Sci. 2007. PMID: 18029420 Free PMC article.
-
An aminoquinazoline inhibitor of the essential bacterial cell wall synthetic enzyme GlmU has a unique non-protein-kinase-like binding mode.Biochem J. 2012 Sep 15;446(3):405-13. doi: 10.1042/BJ20120596. Biochem J. 2012. PMID: 22721802
-
Crystal structures of Streptococcus pneumoniae N-acetylglucosamine-1-phosphate uridyltransferase, GlmU, in apo form at 2.33 A resolution and in complex with UDP-N-acetylglucosamine and Mg(2+) at 1.96 A resolution.J Mol Biol. 2001 Jan 12;305(2):279-89. doi: 10.1006/jmbi.2000.4296. J Mol Biol. 2001. PMID: 11124906
-
Mechanism and Potential Inhibitors of GlmU: A Novel Target for Antimicrobial Drug Discovery.Curr Drug Targets. 2017;18(14):1587-1597. doi: 10.2174/1389450117666160502152011. Curr Drug Targets. 2017. PMID: 27138757 Review.
-
GlmU Inhibitors as Promising Antibacterial Agents: A Review.Mini Rev Med Chem. 2023;23(3):343-360. doi: 10.2174/1389557522666220817114445. Mini Rev Med Chem. 2023. PMID: 35980047 Review.
Cited by
-
Structural and functional features of enzymes of Mycobacterium tuberculosis peptidoglycan biosynthesis as targets for drug development.Tuberculosis (Edinb). 2015 Mar;95(2):95-111. doi: 10.1016/j.tube.2015.01.006. Epub 2015 Jan 29. Tuberculosis (Edinb). 2015. PMID: 25701501 Free PMC article. Review.
-
High-throughput screening identifies novel inhibitors of the acetyltransferase activity of Escherichia coli GlmU.Antimicrob Agents Chemother. 2009 Jun;53(6):2306-11. doi: 10.1128/AAC.01572-08. Epub 2009 Apr 6. Antimicrob Agents Chemother. 2009. PMID: 19349513 Free PMC article.
-
Genetic and structural validation of Aspergillus fumigatus UDP-N-acetylglucosamine pyrophosphorylase as an antifungal target.Mol Microbiol. 2013 Aug;89(3):479-93. doi: 10.1111/mmi.12290. Epub 2013 Jul 5. Mol Microbiol. 2013. PMID: 23750903 Free PMC article.
-
In vitro validation of acetyltransferase activity of GlmU as an antibacterial target in Haemophilus influenzae.J Biol Chem. 2011 Nov 25;286(47):40734-42. doi: 10.1074/jbc.M111.274068. Epub 2011 Oct 7. J Biol Chem. 2011. PMID: 21984832 Free PMC article.
-
GlmU (N-acetylglucosamine-1-phosphate uridyltransferase) bound to three magnesium ions and ATP at the active site.Acta Crystallogr F Struct Biol Commun. 2014 Jun;70(Pt 6):703-8. doi: 10.1107/S2053230X14008279. Epub 2014 May 10. Acta Crystallogr F Struct Biol Commun. 2014. PMID: 24915076 Free PMC article.
References
-
- Anderson, M.S., Raetz, C.R.H. Biosynthesis of lipid A precursors in Escherichia coli. A cytoplasmic acetyltransferase that converts UDP-N-acetylglucosamine to UDP-3-O-(R-3-hydroxymyristoyl)-N-acetylglucosamine. J. Biol. Chem. 1987;262:5159–5169. - PubMed
-
- Brünger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. 1998;D54:905–921. - PubMed
-
- Collaborative Computational Project, Number 4. The CCP4 Suite: Programs for protein crystallography. Acta Crystallogr. 1994;D50:760–763. - PubMed
-
- Gehring, A.M., Lees, W.J., Mindiola, D.J., Walsh, C.T., Brown, E.D. Acetyltransfer precedes uridyltransfer in the formation of UDP-N-acetylglucosamine in separable active sites of the bifunctional glmu protein of Escherichia coli . Biochemistry. 1996;35:579–585. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Medical
