Protein structure and oligomerization are important for the formation of export-competent HIV-1 Rev-RRE complexes
- PMID: 18218716
- PMCID: PMC2248316
- DOI: 10.1110/ps.073246608
Protein structure and oligomerization are important for the formation of export-competent HIV-1 Rev-RRE complexes
Abstract
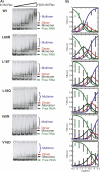
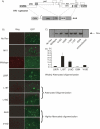
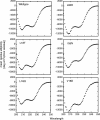
The translation of the unspliced and partially spliced viral mRNAs that encode the late, structural proteins of HIV-1 depends on the viral-protein Rev. Oligomeric binding of Rev to the Rev response element (RRE) in these mRNAs promotes their export from the nucleus and thus controls their expression. Here, we compared the effects of hydrophobic to hydrophilic mutations within the oligomerization domain of Rev using assays for oligomeric RNA binding, protein structure, and export from the nucleus. Oligomeric RNA binding alone does not correlate well with RNA transport activity in the subset of mutants. However, protein structure as judged by CD spectroscopy does correlate well with Rev function. The oligomeric assembly of Rev-L18T is impaired but exhibits minor defects in structure and retains a basal level of activity in vivo. The prevalence of L18T in infected individuals suggests a positive selection mechanism for L18T modulation of Rev activity that may delay the onset of AIDS.
Figures
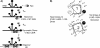


Similar articles
-
Formation of trans-activation competent HIV-1 Rev:RRE complexes requires the recruitment of multiple protein activation domains.PLoS One. 2012;7(6):e38305. doi: 10.1371/journal.pone.0038305. Epub 2012 Jun 4. PLoS One. 2012. PMID: 22675540 Free PMC article.
-
RNA-directed remodeling of the HIV-1 protein Rev orchestrates assembly of the Rev-Rev response element complex.Elife. 2014 Dec 8;3:e04120. doi: 10.7554/eLife.04120. Elife. 2014. PMID: 25486594 Free PMC article.
-
RNA-guided assembly of Rev-RRE nuclear export complexes.Elife. 2014 Aug 27;3:e03656. doi: 10.7554/eLife.03656. Elife. 2014. PMID: 25163983 Free PMC article.
-
HIV Rev Assembly on the Rev Response Element (RRE): A Structural Perspective.Viruses. 2015 Jun 12;7(6):3053-75. doi: 10.3390/v7062760. Viruses. 2015. PMID: 26075509 Free PMC article. Review.
-
[HIV-1 Rev and related inhibitors].Yao Xue Xue Bao. 2007 Apr;42(4):347-51. Yao Xue Xue Bao. 2007. PMID: 17633198 Review. Chinese.
Cited by
-
Formation of trans-activation competent HIV-1 Rev:RRE complexes requires the recruitment of multiple protein activation domains.PLoS One. 2012;7(6):e38305. doi: 10.1371/journal.pone.0038305. Epub 2012 Jun 4. PLoS One. 2012. PMID: 22675540 Free PMC article.
-
Measuring cooperative Rev protein-protein interactions on Rev responsive RNA by fluorescence resonance energy transfer.RNA Biol. 2011 Mar-Apr;8(2):316-24. doi: 10.4161/rna.8.2.13782. Epub 2011 Mar 1. RNA Biol. 2011. PMID: 21358282 Free PMC article.
-
A DEAD-Box Helicase Mediates an RNA Structural Transition in the HIV-1 Rev Response Element.J Mol Biol. 2017 Mar 10;429(5):697-714. doi: 10.1016/j.jmb.2017.01.018. Epub 2017 Jan 31. J Mol Biol. 2017. PMID: 28153748 Free PMC article.
-
Single-molecule studies reveal that DEAD box protein DDX1 promotes oligomerization of HIV-1 Rev on the Rev response element.J Mol Biol. 2011 Jul 29;410(5):959-71. doi: 10.1016/j.jmb.2011.04.026. J Mol Biol. 2011. PMID: 21763499 Free PMC article.
-
HIV-1 Rev-RRE functional activity in primary isolates is highly dependent on minimal context-dependent changes in Rev.Sci Rep. 2022 Nov 1;12(1):18416. doi: 10.1038/s41598-022-21714-2. Sci Rep. 2022. PMID: 36319640 Free PMC article.
References
-
- Andersen, N.H., Liu, Z., Prickett, K.S. Efforts toward deriving the CD spectrum of a 3(10) helix in aqueous medium. FEBS Lett. 1996;399:47–52. - PubMed
-
- Andrade, M.A., Chacon, P., Merelo, J.J., Moran, F. Evaluation of secondary structure of proteins from UV circular dichroism spectra using an unsupervised learning neural network. Protein Eng. 1993;6:383–390. - PubMed
-
- Auer, M., Gremlich, H.U., Seifert, J.M., Daly, T.J., Parslow, T.G., Casari, G., Gstach, H. Helix-loop-helix motif in HIV-1 Rev. Biochemistry. 1994;33:2988–2996. - PubMed
-
- Aukrust, I., Evensen, L., Hollas, H., Berven, F., Atkinson, R.A., Trave, G., Flatmark, T., Vedeler, A. Engineering, biophysical characterisation and binding properties of a soluble mutant form of annexin A2 domain IV that adopts a partially folded conformation. J. Mol. Biol. 2006;363:469–481. - PubMed
-
- Bevec, D., Jaksche, H., Oft, M., Wohl, T., Himmelspach, M., Pacher, A., Schebesta, M., Koettnitz, K., Dobrovnik, M., Csonga, R., et al. Inhibition of HIV-1 replication in lymphocytes by mutants of the Rev cofactor eIF-5A. Science. 1996;271:1858–1860. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
