Conformational pH dependence of intermediate states during oligomerization of the human prion protein
- PMID: 18218718
- PMCID: PMC2248315
- DOI: 10.1110/ps.073163308
Conformational pH dependence of intermediate states during oligomerization of the human prion protein
Abstract
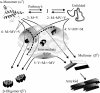
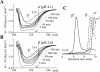
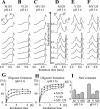
Intermediate states are key to understanding the molecular mechanisms governing protein misfolding. The human prion protein (PrP) can follow various misfolding pathways, and forms a soluble beta-sheet-rich oligomer under acidic, mildly denaturing, high salt conditions. Here we describe a fast conformational switch from the native alpha-monomer to monomeric intermediate states under oligomer-forming conditions, followed by a slower oligomerization process. We observe a pH dependence of the secondary structure of these intermediate forms, with almost native-like alpha-helical secondary structure at pH 4.1 and predominantly beta-sheet characteristics at pH 3.6. NMR spectroscopy differentiates these intermediate states from the native protein and indicates dynamic rearrangements of secondary structure elements characteristic of a molten globule. The alpha-helical intermediate formed at pH 4.1 can convert to the beta-sheet conformation at pH 3.6 but not vice versa, and neither state can be reconverted to an alpha-monomer. The presence of methionine rather than valine at codon 129 accelerates the rate of oligomer formation from the intermediate state.
Figures


Similar articles
-
Oligomerization of the human prion protein proceeds via a molten globule intermediate.J Biol Chem. 2007 Mar 2;282(9):6300-7. doi: 10.1074/jbc.M608926200. Epub 2007 Jan 8. J Biol Chem. 2007. PMID: 17210575
-
NMR characterization of the pH 4 beta-intermediate of the prion protein: the N-terminal half of the protein remains unstructured and retains a high degree of flexibility.Biochem J. 2007 Jan 15;401(2):533-40. doi: 10.1042/BJ20060668. Biochem J. 2007. PMID: 16958619 Free PMC article.
-
The presence of valine at residue 129 in human prion protein accelerates amyloid formation.FEBS Lett. 2005 May 9;579(12):2589-96. doi: 10.1016/j.febslet.2005.03.075. Epub 2005 Apr 8. FEBS Lett. 2005. PMID: 15862295
-
Prion protein oligomer and its neurotoxicity.Acta Biochim Biophys Sin (Shanghai). 2013 Jun;45(6):442-51. doi: 10.1093/abbs/gmt037. Epub 2013 Apr 4. Acta Biochim Biophys Sin (Shanghai). 2013. PMID: 23557632 Review.
-
The pH-triggered conversion of the PrP(c) to PrP(sc.).Curr Top Med Chem. 2013;13(10):1152-63. doi: 10.2174/15680266113139990003. Curr Top Med Chem. 2013. PMID: 23647538 Review.
Cited by
-
Effects of pH and aggregation in the human prion conversion into scrapie form: a study using molecular dynamics with excited normal modes.Eur Biophys J. 2018 Jul;47(5):583-590. doi: 10.1007/s00249-018-1292-4. Epub 2018 Mar 15. Eur Biophys J. 2018. PMID: 29546436
-
Destabilization of polar interactions in the prion protein triggers misfolding and oligomerization.Protein Sci. 2021 Nov;30(11):2258-2271. doi: 10.1002/pro.4188. Epub 2021 Sep 30. Protein Sci. 2021. PMID: 34558139 Free PMC article.
-
Opening Pandora's jar: a primer on the putative roles of CRMP2 in a panoply of neurodegenerative, sensory and motor neuron, and central disorders.Future Neurol. 2012 Nov 1;7(6):749-771. doi: 10.2217/FNL.12.68. Future Neurol. 2012. PMID: 23308041 Free PMC article.
-
Methionine oxidation within the prion protein.Prion. 2020 Dec;14(1):193-205. doi: 10.1080/19336896.2020.1796898. Prion. 2020. PMID: 32744136 Free PMC article. Review.
-
Molecular dynamics simulations capture the misfolding of the bovine prion protein at acidic pH.Biomolecules. 2014 Feb 10;4(1):181-201. doi: 10.3390/biom4010181. Biomolecules. 2014. PMID: 24970211 Free PMC article.
References
-
- Ahmad, A., Millett, I.S., Doniach, S., Uversky, V.N., Fink, A.L. Partially folded intermediates in insulin fibrillation. Biochemistry. 2003;42:11404–11416. - PubMed
-
- Baskakov, I.V., Legname, G., Prusiner, S.B., Cohen, F.E. Folding of prion protein to its native α-helical conformation is under kinetic control. J. Biol. Chem. 2001;276:19687–19690. - PubMed
-
- Baskakov, I.V., Legname, G., Baldwin, M.A., Prusiner, S.B., Cohen, F.E. Pathway complexity of prion protein assembly into amyloid. J. Biol. Chem. 2002;277:21140–21148. - PubMed
-
- Baskakov, I., Disterer, P., Breydo, L., Shaw, M., Gill, A., James, W., Tahiri-Alaoui, A. The presence of valine at residue 129 in human prion protein accelerates amyloid formation. FEBS Lett. 2005;579:2589–2596. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
