Relevance of the flavin binding to the stability and folding of engineered cholesterol oxidase containing noncovalently bound FAD
- PMID: 18218720
- PMCID: PMC2248306
- DOI: 10.1110/ps.073137708
Relevance of the flavin binding to the stability and folding of engineered cholesterol oxidase containing noncovalently bound FAD
Abstract
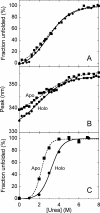
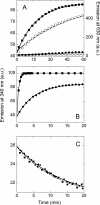
The flavoprotein cholesterol oxidase (CO) from Brevibacterium sterolicum is a monomeric flavoenzyme containing one molecule of FAD cofactor covalently linked to His69. The elimination of the covalent link following the His69Ala substitution was demonstrated to result in a significant decrease in activity, in the midpoint redox potential of the flavin, and in stability with respect to the wild-type enzyme, but does not modify the overall structure of the enzyme. We used CO as a model system to dissect the changes due to the elimination of the covalent link between the flavin and the protein (by comparing the wild-type and H69A CO holoproteins) with those due to the elimination of the cofactor (by comparing the holo- and apoprotein forms of H69A CO). The apoprotein of H69A CO lacks the characteristic tertiary structure of the holoprotein and displays larger hydrophobic surfaces; its urea-induced unfolding does not occur by a simple two-state mechanism and is largely nonreversible. Minor alterations in the flavin binding region are evident between the native and the refolded proteins, and are likely responsible for the low refolding yield observed. A model for the equilibrium unfolding of H69A CO that also takes into consideration the effects of cofactor binding and dissociation, and thus may be of general significance in terms of the relationships between cofactor uptake and folding in flavoproteins, is presented.
Figures






Similar articles
-
Dissecting the structural determinants of the stability of cholesterol oxidase containing covalently bound flavin.J Biol Chem. 2005 Jun 17;280(24):22572-81. doi: 10.1074/jbc.M500549200. Epub 2005 Apr 7. J Biol Chem. 2005. PMID: 15817448
-
Structural and kinetic analyses of the H121A mutant of cholesterol oxidase.Biochem J. 2006 Nov 15;400(1):13-22. doi: 10.1042/BJ20060664. Biochem J. 2006. PMID: 16856877 Free PMC article.
-
Cholesterol oxidase from Brevibacterium sterolicum. The relationship between covalent flavinylation and redox properties.J Biol Chem. 2001 May 25;276(21):18024-30. doi: 10.1074/jbc.M010953200. Epub 2001 Feb 28. J Biol Chem. 2001. PMID: 11359791
-
Dissection of a flavoenzyme active site: the reaction catalyzed by cholesterol oxidase.Antioxid Redox Signal. 2001 Oct;3(5):839-46. doi: 10.1089/15230860152665019. Antioxid Redox Signal. 2001. PMID: 11761331 Review.
-
Cholesterol oxidase: structure and function.Subcell Biochem. 2010;51:137-58. doi: 10.1007/978-90-481-8622-8_5. Subcell Biochem. 2010. PMID: 20213543 Review.
Cited by
-
Contribution of flavin covalent linkage with histidine 99 to the reaction catalyzed by choline oxidase.J Biol Chem. 2009 Jun 19;284(25):16990-16997. doi: 10.1074/jbc.M109.003715. Epub 2009 Apr 27. J Biol Chem. 2009. PMID: 19398559 Free PMC article.
-
Flavin transferase ApbE: From discovery to applications.J Biol Chem. 2025 May;301(5):108453. doi: 10.1016/j.jbc.2025.108453. Epub 2025 Mar 26. J Biol Chem. 2025. PMID: 40154617 Free PMC article. Review.
-
Why the Flavin Adenine Dinucleotide (FAD) Cofactor Needs To Be Covalently Linked to Complex II of the Electron-Transport Chain for the Conversion of FADH2 into FAD.Chemistry. 2018 Apr 6;24(20):5246-5252. doi: 10.1002/chem.201704622. Epub 2017 Dec 14. Chemistry. 2018. PMID: 29124817 Free PMC article.
-
Stabilization of the catalytically active structure of a molybdenum-dependent formate dehydrogenase depends on a highly conserved lysine residue.FEBS J. 2025 Jun;292(12):3165-3179. doi: 10.1111/febs.70048. Epub 2025 Mar 3. FEBS J. 2025. PMID: 40028997 Free PMC article.
-
Fixing Flavins: Hijacking a Flavin Transferase for Equipping Flavoproteins with a Covalent Flavin Cofactor.J Am Chem Soc. 2023 Dec 13;145(49):27140-27148. doi: 10.1021/jacs.3c12009. Epub 2023 Dec 4. J Am Chem Soc. 2023. PMID: 38048072 Free PMC article.
References
-
- Barone, G., Del Vecchio, P., Fessas, D., Giancola, C., Graziano, G. Theseus: A new software package for the handling and analysis of thermal denaturation data of biological macromolecules. J. Therm. Anal. Calimet. 1992;38:2779–2790.
-
- Burstein, E.A., Vedenkina, N.S., Ivkova, M.N. Fluorescence and the location of tryptophan residues in protein molecules. Photochem. Photobiol. 1973;18:263–279. - PubMed
-
- Caldinelli, L., Iametti, S., Barbiroli, A., Bonomi, F., Piubelli, L., Ferranti, P., Picariello, G., Pilone, M.S., Pollegioni, L. Unfolding intermediate in the peroxisomal flavoprotein D-amino acid oxidase. J. Biol. Chem. 2004;279:28426–28434. - PubMed
-
- Caldinelli, L., Iametti, S., Barbiroli, A., Bonomi, F., Fessas, D., Molla, G., Pilone, M.S., Pollegioni, L. Dissecting the structural determinants of the stability of cholesterol oxidase containing covalently bound flavin. J. Biol. Chem. 2005;280:22572–22581. - PubMed
-
- Croteau, N., Vrielink, A. Crystallization and preliminary X-ray analysis of cholesterol oxidase from Brevibacterium sterolicum containing covalently bound FAD. J. Struct. Biol. 1996;116:317–319. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
