The structure of chloroplast DNA molecules and the effects of light on the amount of chloroplast DNA during development in Medicago truncatula
- PMID: 18218970
- PMCID: PMC2259089
- DOI: 10.1104/pp.107.112946
The structure of chloroplast DNA molecules and the effects of light on the amount of chloroplast DNA during development in Medicago truncatula
Abstract
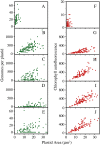
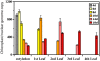
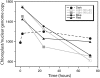
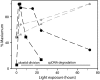
We used pulsed-field gel electrophoresis and restriction fragment mapping to analyze the structure of Medicago truncatula chloroplast DNA (cpDNA). We find most cpDNA in genome-sized linear molecules, head-to-tail genomic concatemers, and complex branched forms with ends at defined sites rather than at random sites as expected from broken circles. Our data suggest that cpDNA replication is initiated predominantly on linear DNA molecules with one of five possible ends serving as putative origins of replication. We also used 4',6-diamidino-2-phenylindole staining of isolated plastids to determine the DNA content per plastid for seedlings grown in the dark for 3 d and then transferred to light before being returned to the dark. The cpDNA content in cotyledons increased after 3 h of light, decreased with 9 h of light, and decreased sharply with 24 h of light. In addition, we used real-time quantitative polymerase chain reaction to determine cpDNA levels of cotyledons in dark- and light-grown (low white, high white, blue, and red light) seedlings, as well as in cotyledons and leaves from plants grown in a greenhouse. In white, blue, and red light, cpDNA increased initially and then declined, but cpDNA declined further in white and blue light while remaining constant in red light. The initial decline in cpDNA occurred more rapidly with increased white light intensity, but the final DNA level was similar to that in less intense light. The patterns of increase and then decrease in cpDNA level during development were similar for cotyledons and leaves. We conclude that the absence in M. truncatula of the prominent inverted repeat cpDNA sequence found in most plant species does not lead to unusual properties with respect to the structure of plastid DNA molecules, cpDNA replication, or the loss of cpDNA during light-stimulated chloroplast development.
Figures

References
-
- Bendich A (1991) Moving pictures of DNA released upon lysis from bacteria, chloroplasts, and mitochondria. Protoplasma 160 121–130
-
- Bendich A, Smith S (1990) Moving pictures and pulsed-field gel electrophoresis show linear DNA molecules from chloroplasts and mitochondria. Curr Genet 17 421–425 - PubMed
-
- Chiu WL, Sears BB (1992) Electron microscopic localization of replication origins in Oenothera chloroplast DNA. Mol Gen Genet 232 33–39 - PubMed
-
- Dubell AN, Mullet JE (1995. a) Continuous far-red light activates plastid DNA synthesis in pea leaves but not full cell enlargement or an increase in plastid number per cell. Plant Physiol 109 95–103
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

