Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion
- PMID: 18218979
- PMCID: PMC2259111
- DOI: 10.1101/gr.7073008
Genome-wide, high-resolution DNA methylation profiling using bisulfite-mediated cytosine conversion
Abstract
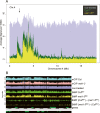
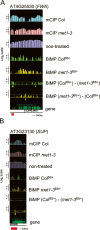
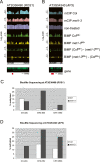
Methylation of cytosines ((m)C) is essential for epigenetic gene regulation in plants and mammals. Aberrant (m)C patterns are associated with heritable developmental abnormalities in plants and with cancer in mammals. We have developed a genome-wide DNA methylation profiling technology employing a novel amplification step for DNA subjected to bisulfite-mediated cytosine conversion. The methylation patterns detected are not only consistent with previous results obtained with (m)C immunoprecipitation (mCIP) techniques, but also demonstrated improved resolution and sensitivity. The technology, named BiMP (for Bisulfite Methylation Profiling), is more cost-effective than mCIP and requires as little as 100 ng of Arabidopsis DNA.
Figures


References
-
- Bao N., Lye K.W., Barton M.K., Lye K.W., Barton M.K., Barton M.K. MicroRNA binding sites in Arabidopsis class III HD-ZIP mRNAs are required for methylation of the template chromosome. Dev. Cell. 2004;7:653–662. - PubMed
-
- Baroux C., Spillane C., Grossniklaus U., Spillane C., Grossniklaus U., Grossniklaus U. Genomic imprinting during seed development. Adv. Genet. 2002;46:165–214. - PubMed
-
- Bird A. DNA methylation patterns and epigenetic memory. Genes & Dev. 2002;16:6–21. - PubMed
-
- Borevitz J.O., Liang D., Plouffe D., Chang H.-S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Liang D., Plouffe D., Chang H.-S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Plouffe D., Chang H.-S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Chang H.-S., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Zhu T., Weigel D., Berry C.C., Winzeler E., Chory J., Weigel D., Berry C.C., Winzeler E., Chory J., Berry C.C., Winzeler E., Chory J., Winzeler E., Chory J., Chory J. Large-scale identification of single-feature polymorphisms in complex genomes. Genome Res. 2003;13:513–523. - PMC - PubMed
-
- Chan S.W., Henderson I.R., Jacobsen S.E., Henderson I.R., Jacobsen S.E., Jacobsen S.E. Gardening the genome: DNA methylation in Arabidopsis thaliana. Nat. Rev. Genet. 2005;6:351–360. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
