BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair
- PMID: 18219312
- PMCID: PMC2267377
- DOI: 10.1038/sj.embor.7401167
BRC-1 acts in the inter-sister pathway of meiotic double-strand break repair
Abstract
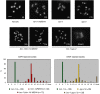
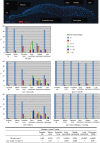
The breast and ovarian cancer susceptibility protein BRCA1 is evolutionarily conserved and functions in DNA double-strand break (DSB) repair through homologous recombination, but its role in meiosis is poorly understood. By using genetic analysis, we investigated the role of the Caenorhabditis elegans BRCA1 orthologue (brc-1) during meiotic prophase. The null mutant in the brc-1 gene is viable, fertile and shows the wild-type complement of six bivalents in most diakinetic nuclei, which is indicative of successful crossover recombination. However, brc-1 mutants show an abnormal increase in apoptosis and RAD-51 foci at pachytene that are abolished by loss of spo-11 function, suggesting a defect in meiosis rather than during premeiotic DNA replication. In genetic backgrounds in which chiasma formation is abrogated, such as him-14/MSH4 and syp-2, loss of brc-1 leads to chromosome fragmentation suggesting that brc-1 is dispensable for crossing over but essential for DSB repair through inter-sister recombination.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
References
-
- Alpi A, Pasierbek P, Gartner A, Loidl J (2003) Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16 - PubMed
-
- Bhattacharyya A, Ear US, Koller BH, Weichselbaum RR, Bishop DK (2000) The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. J Biol Chem 275: 23899–23903 - PubMed
-
- Boulton SJ, Martin JS, Polanowska J, Hill DE, Gartner A, Vidal M (2004) BRCA1/BARD1 orthologs required for DNA repair in Caenorhabditis elegans. Curr Biol 14: 33–39 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

