Histone modifications and chromatin dynamics: a focus on filamentous fungi
- PMID: 18221488
- PMCID: PMC2442719
- DOI: 10.1111/j.1574-6976.2007.00100.x
Histone modifications and chromatin dynamics: a focus on filamentous fungi
Abstract
The readout of the genetic information of eukaryotic organisms is significantly regulated by modifications of DNA and chromatin proteins. Chromatin alterations induce genome-wide and local changes in gene expression and affect a variety of processes in response to internal and external signals during growth, differentiation, development, in metabolic processes, diseases, and abiotic and biotic stresses. This review aims at summarizing the roles of histone H1 and the acetylation and methylation of histones in filamentous fungi and links this knowledge to the huge body of data from other systems. Filamentous fungi show a wide range of morphologies and have developed a complex network of genes that enables them to use a great variety of substrates. This fact, together with the possibility of simple and quick genetic manipulation, highlights these organisms as model systems for the investigation of gene regulation. However, little is still known about regulation at the chromatin level in filamentous fungi. Understanding the role of chromatin in transcriptional regulation would be of utmost importance with respect to the impact of filamentous fungi in human diseases and agriculture. The synthesis of compounds (antibiotics, immunosuppressants, toxins, and compounds with adverse effects) is also likely to be regulated at the chromatin level.
Figures
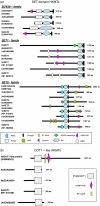


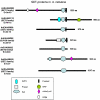
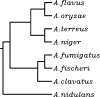


Similar articles
-
[Epigenetic regulation of secondary metabolite biosynthesis in filamentous fungi: a review].Sheng Wu Gong Cheng Xue Bao. 2011 Aug;27(8):1142-8. Sheng Wu Gong Cheng Xue Bao. 2011. PMID: 22097802 Review. Chinese.
-
Epigenomics in stress tolerance of plants under the climate change.Mol Biol Rep. 2023 Jul;50(7):6201-6216. doi: 10.1007/s11033-023-08539-6. Epub 2023 Jun 9. Mol Biol Rep. 2023. PMID: 37294468 Review.
-
The chromatin code of fungal secondary metabolite gene clusters.Appl Microbiol Biotechnol. 2012 Sep;95(6):1389-404. doi: 10.1007/s00253-012-4208-8. Epub 2012 Jul 20. Appl Microbiol Biotechnol. 2012. PMID: 22814413 Free PMC article. Review.
-
H2A.Z and chromatin remodelling complexes: a focus on fungi.Crit Rev Microbiol. 2020 May;46(3):321-337. doi: 10.1080/1040841X.2020.1781784. Epub 2020 Jun 27. Crit Rev Microbiol. 2020. PMID: 32594818 Review.
-
Histone H1 Limits DNA Methylation in Neurospora crassa.G3 (Bethesda). 2016 Jul 7;6(7):1879-89. doi: 10.1534/g3.116.028324. G3 (Bethesda). 2016. PMID: 27172195 Free PMC article.
Cited by
-
Network analysis exposes core functions in major lifestyles of fungal and oomycete plant pathogens.BMC Genomics. 2019 Dec 26;20(1):1020. doi: 10.1186/s12864-019-6409-3. BMC Genomics. 2019. PMID: 31878885 Free PMC article.
-
Histone Methyltransferase SsDim5 Regulates Fungal Virulence through H3K9 Trimethylation in Sclerotinia sclerotiorum.J Fungi (Basel). 2024 Apr 6;10(4):271. doi: 10.3390/jof10040271. J Fungi (Basel). 2024. PMID: 38667942 Free PMC article.
-
Diversity of Fungal DNA Methyltransferases and Their Association With DNA Methylation Patterns.Front Microbiol. 2021 Jan 22;11:616922. doi: 10.3389/fmicb.2020.616922. eCollection 2020. Front Microbiol. 2021. PMID: 33552027 Free PMC article. Review.
-
The Lysine Deacetylase RpdA Is Essential for Virulence in Aspergillus fumigatus.Front Microbiol. 2019 Dec 4;10:2773. doi: 10.3389/fmicb.2019.02773. eCollection 2019. Front Microbiol. 2019. PMID: 31866965 Free PMC article.
-
Fungal Lysine Deacetylases in Virulence, Resistance, and Production of Small Bioactive Compounds.Genes (Basel). 2021 Sep 23;12(10):1470. doi: 10.3390/genes12101470. Genes (Basel). 2021. PMID: 34680865 Free PMC article. Review.
References
-
- Abarca ML, Accensi F, Cano J, Cabanes FJ. Taxonomy and significance of black aspergilli. Antonie Van Leeuwenhoek. 2004;86:33–49. - PubMed
-
- Abe K, Gomi K, Hasegawa F, Machida M. Impact of Aspergillus oryzae genomics on industrial production of metabolites. Mycopathologia. 2006;162:143–153. - PubMed
-
- Adamietz P, Rudolph A. ADP-ribosylation of nuclear proteins in vivo. J Biol Chem. 1984;259:6841–6846. - PubMed
-
- Agalioti T, Chen G, Thanos D. Deciphering the transcriptional histone acetylation code for a human gene. Cell. 2002;111:381–392. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

