Defining functional distances over gene ontology
- PMID: 18221506
- PMCID: PMC2375122
- DOI: 10.1186/1471-2105-9-50
Defining functional distances over gene ontology
Abstract
Background: A fundamental problem when trying to define the functional relationships between proteins is the difficulty in quantifying functional similarities, even when well-structured ontologies exist regarding the activity of proteins (i.e. 'gene ontology' -GO-). However, functional metrics can overcome the problems in the comparing and evaluating functional assignments and predictions. As a reference of proximity, previous approaches to compare GO terms considered linkage in terms of ontology weighted by a probability distribution that balances the non-uniform 'richness' of different parts of the Direct Acyclic Graph. Here, we have followed a different approach to quantify functional similarities between GO terms.
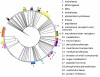
Results: We propose a new method to derive 'functional distances' between GO terms that is based on the simultaneous occurrence of terms in the same set of Interpro entries, instead of relying on the structure of the GO. The coincidence of GO terms reveals natural biological links between the GO functions and defines a distance model Df which fulfils the properties of a Metric Space. The distances obtained in this way can be represented as a hierarchical 'Functional Tree'.
Conclusion: The method proposed provides a new definition of distance that enables the similarity between GO terms to be quantified. Additionally, the 'Functional Tree' defines groups with biological meaning enhancing its utility for protein function comparison and prediction. Finally, this approach could be for function-based protein searches in databases, and for analysing the gene clusters produced by DNA array experiments.
Figures







References
-
- Friedberg I. Automated protein function prediction-the genomic challenge. Brief Bioinform. 2006;7:225–242. - PubMed
-
- Smith B, Kumar A. Controlled vocabularies in bioinformatics: a case study in the gene ontology. DDT: BIOSILICO. 2004;2:246–252.
-
- Rison S, Hodgman T, Thornton J. Comparison of functional annotation schemes for genomes. Funct Integr Genomics. 2000;1:56–69. - PubMed
-
- Valencia A. Automatic annotation of protein function. Current Opinion in Structural Biology. 2005;15:267–74. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

