Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between "open" and "closed" conformations and suggests specific for eEF1A1 affinity for Ca2+-calmodulin
- PMID: 18221514
- PMCID: PMC2275276
- DOI: 10.1186/1472-6807-8-4
Multiple molecular dynamics simulation of the isoforms of human translation elongation factor 1A reveals reversible fluctuations between "open" and "closed" conformations and suggests specific for eEF1A1 affinity for Ca2+-calmodulin
Abstract
Background: Eukaryotic translation elongation factor eEF1A directs the correct aminoacyl-tRNA to ribosomal A-site. In addition, eEF1A is involved in carcinogenesis and apoptosis and can interact with large number of non-translational ligands. There are two isoforms of eEF1A, which are 98% similar. Despite the strong similarity, the isoforms differ in some properties. Importantly, the appearance of eEF1A2 in tissues in which the variant is not normally expressed can be coupled to cancer development.We reasoned that the background for the functional difference of eEF1A1 and eEF1A2 might lie in changes of dynamics of the isoforms.
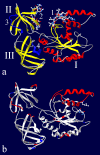
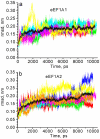
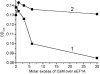
Results: It has been determined by multiple MD simulation that eEF1A1 shows increased reciprocal flexibility of structural domains I and II and less average distance between the domains, while increased non-correlated diffusive atom motions within protein domains characterize eEF1A2. The divergence in the dynamic properties of eEF1A1 and eEF1A2 is caused by interactions of amino acid residues that differ between the two variants with neighboring residues and water environment. The main correlated motion of both protein isoforms is the change in proximity of domains I and II which can lead to disappearance of the gap between the domains and transition of the protein into a "closed" conformation. Such a transition is reversible and the protein can adopt an "open" conformation again. This finding is in line with our earlier experimental observation that the transition between "open" and "closed" conformations of eEF1A could be essential for binding of tRNA and/or other biological ligands. The putative calmodulin-binding region Asn311-Gly327 is less flexible in eEF1A1 implying its increased affinity for calmodulin. The ability of eEF1A1 rather than eEF1A2 to interact with Ca2+/calmodulin is shown experimentally in an ELISA-based test.
Conclusion: We have found that reversible transitions between "open" and "close" conformations of eEF1A provide a molecular background for the earlier observation that the eEF1A molecule is able to change the shape upon interaction with tRNA. The ability of eEF1A1 rather than eEF1A2 to interact with calmodulin is predicted by MD analysis and showed experimentally. The differential ability of the eEF1A isoforms to interact with signaling molecules discovered in this study could be associated with cancer-related properties of eEF1A2.
Figures




Similar articles
-
On the Need to Tell Apart Fraternal Twins eEF1A1 and eEF1A2, and Their Respective Outfits.Int J Mol Sci. 2021 Jun 28;22(13):6973. doi: 10.3390/ijms22136973. Int J Mol Sci. 2021. PMID: 34203525 Free PMC article. Review.
-
Molecular docking uncovers TSPY binds more efficiently with eEF1A2 compared to eEF1A1.J Biomol Struct Dyn. 2015;33(7):1412-23. doi: 10.1080/07391102.2014.952664. Epub 2014 Sep 9. J Biomol Struct Dyn. 2015. PMID: 25105321
-
A2 isoform of mammalian translation factor eEF1A displays increased tyrosine phosphorylation and ability to interact with different signalling molecules.Int J Biochem Cell Biol. 2008;40(1):63-71. doi: 10.1016/j.biocel.2007.08.014. Epub 2007 Sep 1. Int J Biochem Cell Biol. 2008. PMID: 17936057 Free PMC article.
-
Comparison of the ability of mammalian eEF1A1 and its oncogenic variant eEF1A2 to interact with actin and calmodulin.Biol Chem. 2017 Jan 1;398(1):113-124. doi: 10.1515/hsz-2016-0172. Biol Chem. 2017. PMID: 27483363
-
Oncogenic activation of EEF1A2 expression: a journey from a putative to an established oncogene.Cell Mol Biol Lett. 2024 Jan 3;29(1):6. doi: 10.1186/s11658-023-00519-9. Cell Mol Biol Lett. 2024. PMID: 38172654 Free PMC article. Review.
Cited by
-
Structural models of human eEF1A1 and eEF1A2 reveal two distinct surface clusters of sequence variation and potential differences in phosphorylation.PLoS One. 2009 Jul 28;4(7):e6315. doi: 10.1371/journal.pone.0006315. PLoS One. 2009. PMID: 19636410 Free PMC article.
-
On the Need to Tell Apart Fraternal Twins eEF1A1 and eEF1A2, and Their Respective Outfits.Int J Mol Sci. 2021 Jun 28;22(13):6973. doi: 10.3390/ijms22136973. Int J Mol Sci. 2021. PMID: 34203525 Free PMC article. Review.
-
Independent overexpression of the subunits of translation elongation factor complex eEF1H in human lung cancer.BMC Cancer. 2014 Dec 3;14:913. doi: 10.1186/1471-2407-14-913. BMC Cancer. 2014. PMID: 25472873 Free PMC article.
-
Targeting a cluster of arginine residues of neuraminidase to avoid oseltamivir resistance in influenza A (H1N1): a theoretical study.J Mol Model. 2015 Jan;21(1):8. doi: 10.1007/s00894-014-2525-9. Epub 2015 Jan 22. J Mol Model. 2015. PMID: 25605596
-
The eEF1 family of mammalian translation elongation factors.BBA Adv. 2022 Nov 30;3:100067. doi: 10.1016/j.bbadva.2022.100067. eCollection 2023. BBA Adv. 2022. PMID: 37082266 Free PMC article.
References
-
- Negrutskii BS, El'skaya AV. Eukaryotic translation elongation factor 1 alpha: structure, expression, functions, and possible role in aminoacyl-tRNA channeling. Prog Nucleic Acid Res Mol Biol. 1998;60:47–78. - PubMed
-
- Petrushenko ZM, Budkevich TV, Shalak VF, Negrutskii BS, El'skaya AV. Novel complexes of mammalian translation elongation factor eEF1A.GDP with uncharged tRNA and aminoacyl-tRNA synthetase. Implications for tRNA channeling. Eur J Biochem. 2002;269:4811–4818. doi: 10.1046/j.1432-1033.2002.03178.x. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

