The contrasting properties of conservation and correlated phylogeny in protein functional residue prediction
- PMID: 18221517
- PMCID: PMC2267696
- DOI: 10.1186/1471-2105-9-51
The contrasting properties of conservation and correlated phylogeny in protein functional residue prediction
Abstract
Background: Amino acids responsible for structure, core function or specificity may be inferred from multiple protein sequence alignments where a limited set of residue types are tolerated. The rise in available protein sequences continues to increase the power of techniques based on this principle.
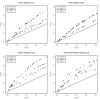
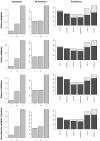
Results: A new algorithm, SMERFS, for predicting protein functional sites from multiple sequences alignments was compared to 14 conservation measures and to the MINER algorithm. Validation was performed on an automatically generated dataset of 1457 families derived from the protein interactions database SNAPPI-DB, and a smaller manually curated set of 148 families. The best performing measure overall was Williamson property entropy, with ROC0.1 scores of 0.0087 and 0.0114 for domain and small molecule contact prediction, respectively. The Lancet method performed worse than random on protein-protein interaction site prediction (ROC0.1 score of 0.0008). The SMERFS algorithm gave similar accuracy to the phylogenetic tree-based MINER algorithm but was superior to Williamson in prediction of non-catalytic transient complex interfaces. SMERFS predicts sites that are significantly more solvent accessible compared to Williamson.
Conclusion: Williamson property entropy is the the best performing of 14 conservation measures examined. The difference in performance of SMERFS relative to Williamson in manually defined complexes was dependent on complex type. The best choice of analysis method is therefore dependent on the system of interest. Additional computation employed by Miner in calculation of phylogenetic trees did not produce improved results over SMERFS. SMERFS performance was improved by use of windows over alignment columns, illustrating the necessity of considering the local environment of positions when assessing their functional significance.
Figures




References
-
- Genome Pages at the EBI http://www.ebi.ac.uk/genomes/
-
- Do JH, Choi DK. Computational approaches to gene prediction. J Microbiol. 2006;44:137–144. - PubMed
-
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. - DOI - PMC - PubMed
-
- Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. SDPpred: a tool for prediction of amino acid residues thatdetermine differences in functional specificity of homologousproteins. Nucleic Acids Res. 2004:W424–8. doi: 10.1093/nar/gkh391. [1362-4962 Journal Article]. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

