The development of PIPA: an integrated and automated pipeline for genome-wide protein function annotation
- PMID: 18221520
- PMCID: PMC2259298
- DOI: 10.1186/1471-2105-9-52
The development of PIPA: an integrated and automated pipeline for genome-wide protein function annotation
Abstract
Background: Automated protein function prediction methods are needed to keep pace with high-throughput sequencing. With the existence of many programs and databases for inferring different protein functions, a pipeline that properly integrates these resources will benefit from the advantages of each method. However, integrated systems usually do not provide mechanisms to generate customized databases to predict particular protein functions. Here, we describe a tool termed PIPA (Pipeline for Protein Annotation) that has these capabilities.
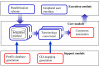
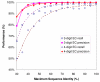
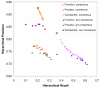
Results: PIPA annotates protein functions by combining the results of multiple programs and databases, such as InterPro and the Conserved Domains Database, into common Gene Ontology (GO) terms. The major algorithms implemented in PIPA are: (1) a profile database generation algorithm, which generates customized profile databases to predict particular protein functions, (2) an automated ontology mapping generation algorithm, which maps various classification schemes into GO, and (3) a consensus algorithm to reconcile annotations from the integrated programs and databases.PIPA's profile generation algorithm is employed to construct the enzyme profile database CatFam, which predicts catalytic functions described by Enzyme Commission (EC) numbers. Validation tests show that CatFam yields average recall and precision larger than 95.0%. CatFam is integrated with PIPA. We use an association rule mining algorithm to automatically generate mappings between terms of two ontologies from annotated sample proteins. Incorporating the ontologies' hierarchical topology into the algorithm increases the number of generated mappings. In particular, it generates 40.0% additional mappings from the Clusters of Orthologous Groups (COG) to EC numbers and a six-fold increase in mappings from COG to GO terms. The mappings to EC numbers show a very high precision (99.8%) and recall (96.6%), while the mappings to GO terms show moderate precision (80.0%) and low recall (33.0%). Our consensus algorithm for GO annotation is based on the computation and propagation of likelihood scores associated with GO terms. The test results suggest that, for a given recall, the application of the consensus algorithm yields higher precision than when consensus is not used.
Conclusion: The algorithms implemented in PIPA provide automated genome-wide protein function annotation based on reconciled predictions from multiple resources.
Figures

Similar articles
-
Genome-wide enzyme annotation with precision control: catalytic families (CatFam) databases.Proteins. 2009 Feb 1;74(2):449-60. doi: 10.1002/prot.22167. Proteins. 2009. PMID: 18636476
-
ProLoc-GO: utilizing informative Gene Ontology terms for sequence-based prediction of protein subcellular localization.BMC Bioinformatics. 2008 Feb 1;9:80. doi: 10.1186/1471-2105-9-80. BMC Bioinformatics. 2008. PMID: 18241343 Free PMC article.
-
An evaluation of GO annotation retrieval for BioCreAtIvE and GOA.BMC Bioinformatics. 2005;6 Suppl 1(Suppl 1):S17. doi: 10.1186/1471-2105-6-S1-S17. Epub 2005 May 24. BMC Bioinformatics. 2005. PMID: 15960829 Free PMC article.
-
How to learn about gene function: text-mining or ontologies?Methods. 2015 Mar;74:3-15. doi: 10.1016/j.ymeth.2014.07.004. Epub 2014 Aug 1. Methods. 2015. PMID: 25088781 Review.
-
In silico characterization of proteins: UniProt, InterPro and Integr8.Mol Biotechnol. 2008 Feb;38(2):165-77. doi: 10.1007/s12033-007-9003-x. Epub 2007 Oct 4. Mol Biotechnol. 2008. PMID: 18219596 Review.
Cited by
-
Identification and optimization of classifier genes from multi-class earthworm microarray dataset.PLoS One. 2010 Oct 28;5(10):e13715. doi: 10.1371/journal.pone.0013715. PLoS One. 2010. PMID: 21060837 Free PMC article.
-
The automatic annotation of bacterial genomes.Brief Bioinform. 2013 Jan;14(1):1-12. doi: 10.1093/bib/bbs007. Epub 2012 Mar 9. Brief Bioinform. 2013. PMID: 22408191 Free PMC article.
-
AGeS: a software system for microbial genome sequence annotation.PLoS One. 2011 Mar 7;6(3):e17469. doi: 10.1371/journal.pone.0017469. PLoS One. 2011. PMID: 21408217 Free PMC article.
-
Quantitative frame analysis and the annotation of GC-rich (and other) prokaryotic genomes. An application to Anaeromyxobacter dehalogenans.Bioinformatics. 2015 Oct 15;31(20):3254-61. doi: 10.1093/bioinformatics/btv339. Epub 2015 Jun 4. Bioinformatics. 2015. PMID: 26048600 Free PMC article.
-
Integration of bioinformatics to biodegradation.Biol Proced Online. 2014 Apr 27;16:8. doi: 10.1186/1480-9222-16-8. eCollection 2014. Biol Proced Online. 2014. PMID: 24808763 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

