Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses
- PMID: 18221523
- PMCID: PMC2262068
- DOI: 10.1186/1471-2164-9-42
Probing regulon of ArcA in Shewanella oneidensis MR-1 by integrated genomic analyses
Abstract
Background: The Arc two-component system is a global regulator controlling many genes involved in aerobic/anaerobic respiration and fermentative metabolism in Escherichia coli. Shewanella oneidensis MR-1 contains a gene encoding a putative ArcA homolog with ~81% amino acid sequence identity to the E. coli ArcA protein but not a full-length arcB gene.
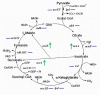
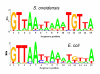
Results: To understand the role of ArcA in S. oneidensis, an arcA deletion strain was constructed and subjected to both physiological characterization and microarray analysis. Compared to the wild-type MR-1, the mutant exhibited impaired aerobic growth and a defect in utilizing DMSO in the absence of O2. Microarray analyses on cells grown aerobically and anaerobically on fumarate revealed that expression of 1009 genes was significantly affected (p < 0.05) by the mutation. In contrast to E. coli ArcA, the protein appears to be dispensable in regulation of the TCA cycle in S. oneidensis. To further determine genes regulated by the Arc system, an ArcA recognition weight matrix from DNA-binding data and bioinformatics analysis was generated and used to produce an ArcA sequence affinity map. By combining both techniques, we identified an ArcA regulon of at least 50 operons, of which only 6 were found to be directly controlled by ArcA in E. coli.
Conclusion: These results indicate that the Arc system in S. oneidensis differs from that in E. coli substantially in terms of its physiological function and regulon while their binding motif are strikingly similar.
Figures





Similar articles
-
Anaerobic regulation by an atypical Arc system in Shewanella oneidensis.Mol Microbiol. 2005 Jun;56(5):1347-57. doi: 10.1111/j.1365-2958.2005.04628.x. Mol Microbiol. 2005. PMID: 15882425
-
Physiological roles of ArcA, Crp, and EtrA and their interactive control on aerobic and anaerobic respiration in Shewanella oneidensis.PLoS One. 2010 Dec 28;5(12):e15295. doi: 10.1371/journal.pone.0015295. PLoS One. 2010. PMID: 21203399 Free PMC article.
-
Impaired cell envelope resulting from arcA mutation largely accounts for enhanced sensitivity to hydrogen peroxide in Shewanella oneidensis.Sci Rep. 2015 May 15;5:10228. doi: 10.1038/srep10228. Sci Rep. 2015. PMID: 25975178 Free PMC article.
-
Cellular and molecular physiology of Escherichia coli in the adaptation to aerobic environments.J Biochem. 1996 Dec;120(6):1055-63. doi: 10.1093/oxfordjournals.jbchem.a021519. J Biochem. 1996. PMID: 9010748 Review.
-
Aerobic-anaerobic gene regulation in Escherichia coli: control by the ArcAB and Fnr regulons.Res Microbiol. 1994 Jun-Aug;145(5-6):437-50. doi: 10.1016/0923-2508(94)90092-2. Res Microbiol. 1994. PMID: 7855430 Review.
Cited by
-
The transcription factors ActR and SoxR differentially affect the phenazine tolerance of Agrobacterium tumefaciens.Mol Microbiol. 2019 Jul;112(1):199-218. doi: 10.1111/mmi.14263. Epub 2019 May 3. Mol Microbiol. 2019. PMID: 31001852 Free PMC article.
-
Genetic and molecular characterization of flagellar assembly in Shewanella oneidensis.PLoS One. 2011;6(6):e21479. doi: 10.1371/journal.pone.0021479. Epub 2011 Jun 22. PLoS One. 2011. PMID: 21731763 Free PMC article.
-
Exoprotein production correlates with morphotype changes of nonmotile Shewanella oneidensis mutants.J Bacteriol. 2013 Apr;195(7):1463-74. doi: 10.1128/JB.02187-12. Epub 2013 Jan 18. J Bacteriol. 2013. PMID: 23335418 Free PMC article.
-
Electrochemically active bacteria sense electrode potentials for regulating catabolic pathways.Nat Commun. 2018 Mar 14;9(1):1083. doi: 10.1038/s41467-018-03416-4. Nat Commun. 2018. PMID: 29540717 Free PMC article.
-
Protection from oxidative stress relies mainly on derepression of OxyR-dependent KatB and Dps in Shewanella oneidensis.J Bacteriol. 2014 Jan;196(2):445-58. doi: 10.1128/JB.01077-13. Epub 2013 Nov 8. J Bacteriol. 2014. PMID: 24214945 Free PMC article.
References
-
- Nealson KH, Scott J. Ecophysiology of the genus Shewanella. In: Dworkin M, editor. The Prokaryotes. Springer-Verlag; 2004.
-
- Heidelberg JF, Paulsen IT, Nelson KE, Gaidos EJ, Nelson WC, Read TD, Eisen JA, Seshadri R, Ward N, Methe B, Clayton RA, Meyer T, Tsapin A, Scott J, Beanan M, Brinkac L, Daugherty S, DeBoy RT, Dodson RJ, Durkin AS, Haft DH, Kolonay JF, Madupu R, Peterson JD, Umayam LA, White O, Wolf AM, Vamathevan J, Weidman J, Impraim M, Lee K, Berry K, Lee C, Mueller J, Khouri H, Gill J, Utterback TR, McDonald LA, Feldblyum TV, Smith HO, Venter JC, Nealson KH, Fraser CM. Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis. Nat Biotech. 2002;20:1118–1123. doi: 10.1038/nbt749. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

